Worm Development
| Embryology - 25 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction
Initially used in the 1960's by Sydney Brenner to study the genetics of development and neurobiology. Early embryological studies of the nematode worm (roundworm) Caenorhabditis elegans (C.Elegans, so called because of its "elegant" curving movement) characterized the fate of each and every cell in the worm through all stages of development. This worm was the first to have its entire genome sequenced and also used recently in space experiments (see below).
- Sydney Brenner died on April 5, 2019, at age 92.[1]
The USA space shuttle Atlantis in November 2009 launched Caenorhabditis elegans into space as part of an experiment to study RNA interference and protein phosphorylation in a space environment.
- "RNA interference and protein phosphorylation in space environment using the nematode Caenorhabditis elegans (CERISE) is an experiment that addresses two scientific objectives. The first is to evaluate the effect of microgravity on ribonucleic acid (RNA) interference. The second is to study how the space environment effects protein phosphorylation (addition of a phosphate molecule) and signal transduction in the muscle fibers of gene knock-downed Caenorhabditis elegans."
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Caenorhabditis elegans Development | Worm Development | Worm Embryology |
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
Adult Anatomy
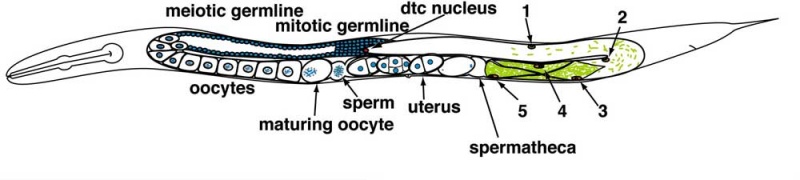
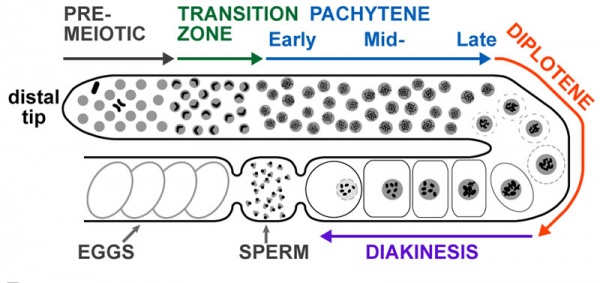
Adult Hermaphrodite Gonad
Adult hermaphrodite gonad arm[8] - A drawing representation of an adult hermaphrodite gonad arm. The progression of germ cell proliferation and meiosis are indicated by the arrows starting from the distal tip region of the gonad arm.
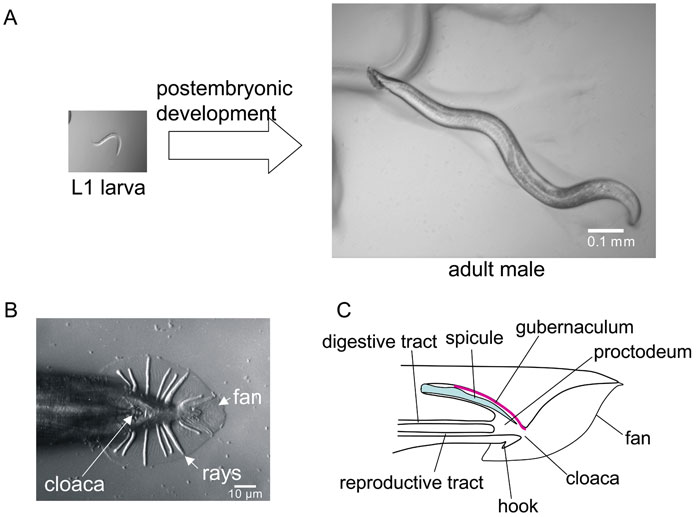
Male Development
The features that differentiate the C. elegans male from the hermaphrodite arise during postembryonic development.[9]
RNA interference
The two researchers, Andrew Z. Fire and Craig C. Mello[10], were investigating how gene expression is regulated in C. elegans and identified the novel regulation method of RNA interference (RNAi), gene silencing by double-stranded RNA. This discovery was awarded the 2006 Nobel Prize in Physiology or Medicine.
- Links: 2006 Nobel Press Release
Embryonic Cell Lineages
The overview diagram above shows the fate of each individual cell in the developing c. elegans.
- Zygote (P0 cell) divides into two daughter cells (AB and P1 cells).
- These two daughter cells then divide into the next generation.
- the "X" indicates cells that die by apoptosis during development.
Note the above image is not at a readable resolution, to view see large readable version (10,389 × 1,336 pixels). Embryonic cell lineage developed by J .E. Sulston, E. Schierenberg, J. G. White, J. N. Thomson.
- Links: Apoptosis | Worm Atlas - Cell Lineages
Gastrointestinal Tract
The worm digestive tract consists of a pharynx (80-cell), intestine, and rectum and contains only about 100 cells. Development is regulated by similar transcription factors found for other species (FoxA and GATA factors).[11] There are also 20 neurons located within the pharynx.{#pmid:15780187|PMID15780187}}
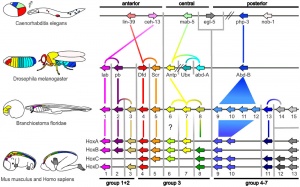
Developmental Genes
- pha-4 (Drosophila forkhead and vertebrate FoxA) - pharynx and rectum
- ceh-22 (Drosophila tinman and vertebrate Template:Nkx2.5) - pharynx
- pha-2 (vertebrate Hex) - pharynx
- GATA - intestine
References
- ↑ Pederson T. (2019). The sui generis Sydney Brenner. Proc. Natl. Acad. Sci. U.S.A. , 116, 13155-13157. PMID: 31182578 DOI.
- ↑ Hueber SD, Weiller GF, Djordjevic MA & Frickey T. (2010). Improving Hox protein classification across the major model organisms. PLoS ONE , 5, e10820. PMID: 20520839 DOI.
- ↑ Reilly MB, Cros C, Varol E, Yemini E & Hobert O. (2020). Unique homeobox codes delineate all the neuron classes of C. elegans. Nature , , . PMID: 32814896 DOI.
- ↑ Kurland M, O'Meara B, Tucker DK & Ackley BD. (2020). The Hox Gene egl-5 Acts as a Terminal Selector for VD13 Development via Wnt Signaling. J Dev Biol , 8, . PMID: 32138237 DOI.
- ↑ Hench J, Henriksson J, Abou-Zied AM, Lüppert M, Dethlefsen J, Mukherjee K, Tong YG, Tang L, Gangishetti U, Baillie DL & Bürglin TR. (2015). The Homeobox Genes of Caenorhabditis elegans and Insights into Their Spatio-Temporal Expression Dynamics during Embryogenesis. PLoS ONE , 10, e0126947. PMID: 26024448 DOI.
- ↑ Porta-de-la-Riva M, Fontrodona L, Villanueva A & Cerón J. (2012). Basic Caenorhabditis elegans methods: synchronization and observation. J Vis Exp , , e4019. PMID: 22710399 DOI.
- ↑ Resnick TD, McCulloch KA & Rougvie AE. (2010). miRNAs give worms the time of their lives: small RNAs and temporal control in Caenorhabditis elegans. Dev. Dyn. , 239, 1477-89. PMID: 20232378 DOI.
- ↑ Bickel JS, Chen L, Hayward J, Yeap SL, Alkers AE & Chan RC. (2010). Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. , 6, e1001028. PMID: 20661436 DOI.
- ↑ Emmons SW. (2005). Male development. WormBook , , 1-22. PMID: 18050419 DOI.
- ↑ Timmons L, Tabara H, Mello CC & Fire AZ. (2003). Inducible systemic RNA silencing in Caenorhabditis elegans. Mol. Biol. Cell , 14, 2972-83. PMID: 12857879 DOI.
- ↑ Kormish JD, Gaudet J & McGhee JD. (2010). Development of the C. elegans digestive tract. Curr. Opin. Genet. Dev. , 20, 346-54. PMID: 20570129 DOI.
Books
WormBook - a comprehensive, open-access collection of original, peer-reviewed chapters covering topics related to the biology of Caenorhabditis elegans and other nematodes.
Nigon VM & Félix MA. (2017). History of research on C. elegans and other free-living nematodes as model organisms. WormBook , 2017, 1-84. PMID: 28326696 DOI.
Gieseler K, Qadota H & Benian GM. (2017). Development, structure, and maintenance of C. elegans body wall muscle. WormBook , 2017, 1-59. PMID: 27555356 DOI.
Hillers KJ, Jantsch V, Martinez-Perez E & Yanowitz JL. (2017). Meiosis. WormBook , 2017, 1-43. PMID: 26694509 DOI.
- Asymmetric cell division and axis formation in the embryo
- Embryological variation during nematode development
- Links: Search WormBook by date
Reviews
Liang JJH, McKinnon IA & Rankin CH. (2020). The contribution of C. elegans neurogenetics to understanding neurodegenerative diseases. J. Neurogenet. , , 1-22. PMID: 32772603 DOI.
Rothman J & Jarriault S. (2019). Developmental Plasticity and Cellular Reprogramming in Caenorhabditis elegans. Genetics , 213, 723-757. PMID: 31685551 DOI.
Dimov I & Maduro MF. (2019). The C. elegans intestine: organogenesis, digestion, and physiology. Cell Tissue Res. , 377, 383-396. PMID: 31065800 DOI.
Pintard L & Bowerman B. (2019). Mitotic Cell Division in Caenorhabditis elegans. Genetics , 211, 35-73. PMID: 30626640 DOI.
Murray JI. (2018). Systems biology of embryonic development: Prospects for a complete understanding of the Caenorhabditis elegans embryo. Wiley Interdiscip Rev Dev Biol , 7, e314. PMID: 29369536 DOI.
Maduro MF. (2017). Gut development in C. elegans. Semin. Cell Dev. Biol. , 66, 3-11. PMID: 28065852 DOI.
Articles
Rödelsperger C, Ebbing A, Sharma DR, Okumura M, Sommer RJ & Korswagen HC. (2020). Spatial transcriptomics of nematodes identifies sperm cells as a source of genomic novelty and rapid evolution. Mol. Biol. Evol. , , . PMID: 32785688 DOI.
Search Pubmed
July 2010 "c elegans Development" All (5126) Review (898) Free Full Text (2363)
Search Pubmed: Worm Development | Caenorhabditis elegans Development | c elegans Development
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- WormBook open-access collection of chapters covering topics related to the biology of Caenorhabditis elegans (C. elegans) and other nematodes.
- Caenorhabditis Genome Sequencing Projects
- Caenorhabditis elegans WWW Server
- The Hall lab - aim to serve the C. elegans community by creating a digital database of TEM images & wild type tissues- an Atlas of worm anatomy.
| Animal Development: axolotl | bat | cat | chicken | cow | dog | dolphin | echidna | fly | frog | goat | grasshopper | guinea pig | hamster | horse | kangaroo | koala | lizard | medaka | mouse | opossum | pig | platypus | rabbit | rat | salamander | sea squirt | sea urchin | sheep | worm | zebrafish | life cycles | development timetable | development models | K12 |
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 25) Embryology Worm Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Worm_Development
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G