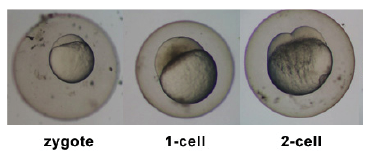
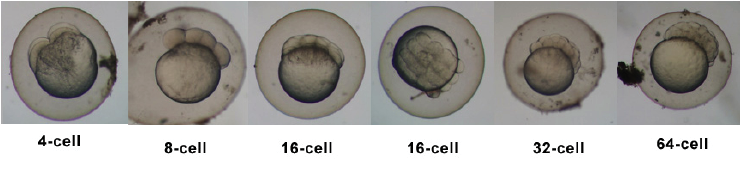
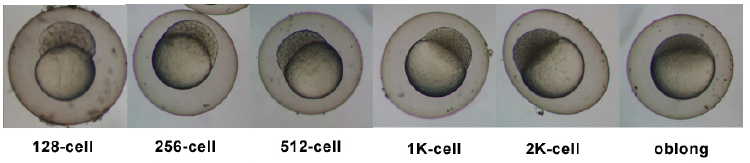
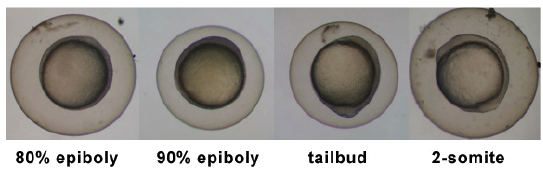
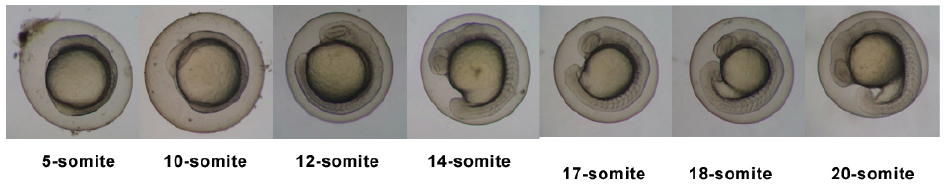
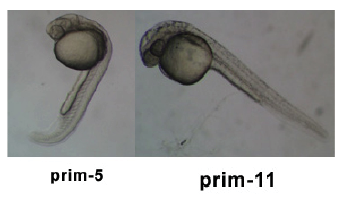
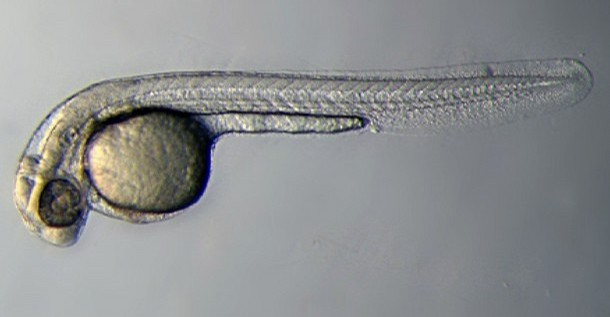
Introduction
Zebrafish or zebra danio (danio rerio) are seen as one of the latest "models" for vertebrate embryological development studies. These embryos have the great advantage that they develop as "see through" embryos, that is, all internal development can be clearly observed from the outside in the living embryo. Much of the early modern work using this embryo model began with the papers of Kimmel.[1][2]
Several large laboratories in the US are now developing large breeding programs to carry out "knockouts" and to find spontaneous mutants of interest.
Some Recent Findings
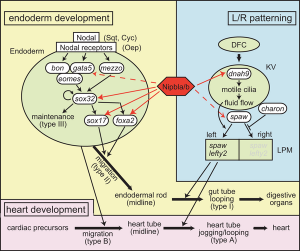
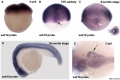
Nipbl heart and organ patterning
[3]
- Novel Development of Magnetic Resonance Imaging to Quantify the Structural Anatomic Growth of Diverse Organs in Adult and Mutant Zebrafish[4] "Zebrafish (Danio rerio) is a widely used vertebrate animal for modeling genetic diseases by targeted editing strategies followed by gross phenotypic and biomarker characterization. While larval transparency permits microscopic detection of anatomical defects, histological adult screening for organ-level defects remains invasive, tedious, inefficient, and subject to technical artifact. Here, we describe a noninvasive magnetic resonance imaging (MRI) approach to systematically screen adult zebrafish for anatomical growth defects."
- ZebraShare: a new venue for rapid dissemination of zebrafish mutant data[5] "Background: In the past decade, the zebrafish community has widely embraced targeted mutagenesis technologies, resulting in an abundance of mutant lines. While many lines have proven to be useful for investigating gene function, many have also shown no apparent phenotype, or phenotypes not of interest to the originating lab. In order for labs to document and share information about these lines, we have created ZebraShare as a new resource offered within ZFIN. ZebraShare involves a form-based submission process generated by ZFIN. The ZebraShare interface (https://zfin.org/action/zebrashare) can be accessed on ZFIN under "Submit Data".
- Involvement of homeobox transcription factor Mohawk in palatogenesis[6] "Palatogenesis is affected by many factors, including gene polymorphisms and exposure to toxic chemicals during sensitive developmental periods. Cleft palate is one of the most common congenital anomalies, and ongoing efforts to elucidate the molecular mechanisms underlying palatogenesis are providing useful insights to reduce the risk of this disorder. To identify novel potential regulators of palatogenesis, we analyzed public transcriptome datasets from a mouse model of cleft palate caused by selective deletion of transforming growth factor-β (TGFβ) receptor type 2 in cranial neural crest cells. We identified the homeobox transcription factor Mohawk (Mkx) as a gene downregulated in the maxilla of TGFβ knockout mice compared with wild-type mice. To examine the role of mkx in palatogenesis, we used CRISPR/Cas9 editing to generate zebrafish with impaired expression of mkxa and mkxb, the zebrafish homologs of Mkx. We found that mkx crispants expressed reduced levels of gli1, a critical transcription factor in the Sonic hedgehog (SHH) signaling pathway that plays an important role in the regulation of palatogenesis."
- Conserved Genoarchitecture of the Basal Hypothalamus in Zebrafish Embryos[7] "Analyses of genoarchitecture recently stimulated substantial revisions of anatomical models for the developing hypothalamus in mammalian and other vertebrate systems. The prosomeric model proposes the hypothalamus to be derived from the secondary prosencephalon, and to consist of alar and basal regions. The basal hypothalamus can further be subdivided into tuberal and mamillary regions, each with distinct subregions. Albeit being a widely used model system for neurodevelopmental studies, no detailed genoarchitectural maps exist for the zebrafish (Danio rerio) hypothalamus. Here, we compare expression domains of zebrafish genes, including arxa, Shha, otpa, isl1, lhx5, nkx2.1, nkx2.2a, pax6, and dlx5a, the orthologs of which delimit specific subregions within the murine basal hypothalamus. We develop the highly conserved brain-specific homeobox (bsx) gene as a novel marker for genoarchitectural analysis of hypothalamic regions. Our comparison of gene expression patterns reveals that the genoarchitecture of the basal hypothalamus in zebrafish embryos 48 hours post fertilization is highly similar to mouse embryos at E13.5. We found the tuberal hypothalamus in zebrafish embryos to be relatively large and to comprise previously ill-defined regions around the posterior hypothalamic recess. The mamillary hypothalamus is smaller and concentrates to rather medial areas in proximity to the anterior end of the neural tube floor plate. Within the basal hypothalamus we identified longitudinal and transverse tuberal and mamillary subregions topologically equivalent to those previously described in other vertebrates. However, the hypothalamic diencephalic boundary region and the posterior tuberculum still provide a challenge. We applied the updated prosomeric model to the developing zebrafish hypothalamus to facilitate cross-species comparisons. Accordingly, we applied the mammalian nomenclature of hypothalamic organization to zebrafish and propose it to replace some controversial previous nomenclature."
|
| More recent papers
|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
- This search now requires a manual link as the original PubMed extension has been disabled.
- The displayed list of references do not reflect any editorial selection of material based on content or relevance.
- References also appear on this list based upon the date of the actual page viewing.
References listed on the rest of the content page and the associated discussion page (listed under the publication year sub-headings) do include some editorial selection based upon both relevance and availability.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References
Search term: Zebrafish Embryology | Zebrafish Development
|
| Older papers
|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
- Review - Zebrafish as a model for studying ovarian development: Recent advances from targeted gene knockout studies[8] "Ovarian development is a complex process controlled by precise coordination of multiple factors. The targeted gene knockout technique is a powerful tool to study the functions of these factors. The successful application of this technique in mice in the past three decades has significantly enhanced our understanding on the molecular mechanism of ovarian development. Recently, with the advent of genome editing techniques, targeted gene knockout research can be carried out in many species. Zebrafish has emerged as an excellent model system to study the control of ovarian development. Dozens of genes related to ovarian development have been knocked out in zebrafish in recent years. Much new information and perspectives on the molecular mechanism of ovarian development have been obtained from these mutant zebrafish. Some findings have challenged conventional views. Several genes have been identified for the first time in vertebrates to control ovarian development. Focusing on ovarian development, the purpose of this review is to briefly summarize recent findings using these gene knockout zebrafish models, and compare these findings with mammalian models." ovary
- Anatomy, development, and plasticity of the neurosecretory hypothalamus in zebrafish[9] "The paraventricular nucleus (PVN) of the hypothalamus harbors diverse neurosecretory cells with critical physiological roles for the homeostasis. Decades of research in rodents have provided a large amount of information on the anatomy, development, and function of this important hypothalamic nucleus. However, since the hypothalamus lies deep within the brain in mammals and is difficult to access, many questions regarding development and plasticity of this nucleus still remain. In particular, how different environmental conditions, including stress exposure, shape the development of this important nucleus has been difficult to address in animals that develop in utero. To address these open questions, the transparent larval zebrafish with its rapid external development and excellent genetic toolbox offers exciting opportunities. In this review, we summarize recent information on the anatomy and development of the neurosecretory preoptic area (NPO), which represents a similar structure to the mammalian PVN in zebrafish. We will then review recent studies on the development of different cell types in the neurosecretory hypothalamus both in mouse and in fish. Lastly, we discuss stress-induced plasticity of the PVN mainly discussing the data obtained in rodents, but pointing out tools and approaches available in zebrafish for future studies. This review serves as a primer for the currently available information relevant for studying the development and plasticity of this important brain region using zebrafish." hypothalamus
- Review - Development cell by cell[10] "The result is the ability to track development of organisms and organs in stunning detail, cell by cell and through time. Science is recognizing that combination of technologies, and its potential for spurring advances in basic research and medicine, as the 2018 Breakthrough of the Year."
- Zebrafish znfl1s regulate left-right asymmetry patterning through controlling the expression of fgfr1a[11] "Proper left-right (LR) axis establishment is critical for organogenesis in vertebrates. Previously, we reported that zinc finger transcription factors zinc finger transcription factor 1 (znfl1s) are expressed in the tailbud and axial mesoderm in zebrafish. However, a role of znfl1s in LR axis development has not been demonstrated. Here, we discovered that the knockdown of znfl1s using morpholino (MO) in whole embryos or dorsal forerunner cells (DFCs) interrupted LR asymmetry and normal development of the heart, liver, and pancreas. Whole-embryo knockdown of znfl1s by MO or clustered regularly interspaced short palindromic repeat (CRISPR) interference (CRISPRi) resulted in the absent expression of nodal gene spaw and Nodal signaling-related genes lft1, lft2, and pitx2c in the left lateral plate mesoderm (LPM), and Spaw, Lft1, Lft2, and Pitx2c play important roles in LR axis development in zebrafish. However, specific knockdown of znfl1s in DFCs resulted in random expression of spaw, lft1, lft2, and pitx2c. Knockdown of znfl1s led to abnormal cilia formation by the downregulation of fgfr1a and foxj1a expression. The expression of spaw, lft1, lft2, and pitx2c was partially rescued by the overexpression of fgfr1a mRNA in znfl1s morphants. Taken together, our results suggest that znfl1s regulate laterality development in zebrafish embryos through controlling the expression of fgfr1a."
- Zebrafish Pronephros Development[12] "The pronephros is the first kidney type to form in vertebrate embryos. The first step of pronephrogenesis in the zebrafish is the formation of the intermediate mesoderm during gastrulation, which occurs in response to secreted morphogens such as BMPs and Nodals. Patterning of the intermediate mesoderm into proximal and distal cell fates is induced by retinoic acid signaling with downstream transcription factors including wt1a, pax2a, pax8, hnf1b, sim1a, mecom, and irx3b. In the anterior intermediate mesoderm, progenitors of the glomerular blood filter migrate and fuse at the midline and recruit a blood supply. More posteriorly localized tubule progenitors undergo epithelialization and fuse with the cloaca. The Notch signaling pathway regulates the formation of multi-ciliated cells in the tubules and these cells help propel the filtrate to the cloaca. The lumenal sheer stress caused by flow down the tubule activates anterior collective migration of the proximal tubules and induces stretching and proliferation of the more distal segments. Ultimately these processes create a simple two-nephron kidney that is capable of reabsorbing and secreting solutes and expelling excess water-processes that are critical to the homeostasis of the body fluids. The zebrafish pronephric kidney provides a simple, yet powerful, model system to better understand the conserved molecular and cellular progresses that drive nephron formation, structure, and function." renal
- A crystal-clear zebrafish for in vivo imaging[13] "Here we present crystal, an optically clear zebrafish mutant obtained by combining different viable mutations affecting skin pigmentation. Compared to the previously described combinatorial mutant casper, the crystal mutant lacks pigmentation also in the retinal pigment epithelium, therefore enabling optical access to the eyes. Unlike PTU-treated animals, crystal larvae are able to perform visually guided behaviours, such as the optomotor response, as efficiently as wild type larvae. To validate the in vivo application of crystal larvae, we performed whole-brain light-sheet imaging and two-photon calcium imaging of neural activity in the retina."
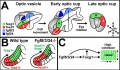
- Construction of a vertebrate embryo from two opposing morphogen gradients[14] "Here, we show that opposing gradients of bone morphogenetic protein (BMP) and Nodal, two transforming growth factor family members that act as morphogens, are sufficient to induce molecular and cellular mechanisms required to organize, in vivo or in vitro, uncommitted cells of the zebrafish blastula animal pole into a well-developed embryo." BMP
- FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution[15] "We present the Fish Face Atlas, an online, 3D-interactive atlas of craniofacial development in the zebrafish Danio rerio. Alizarin red-stained skulls scanned by fluorescent optical projection tomography and segmented into individual elements provide a resource for understanding the 3D structure of the zebrafish craniofacial skeleton." See also Online Zebrafish Atlases
- Multifactorial Origins of Heart and Gut Defects in nipbl-Deficient Zebrafish, a Model of Cornelia de Lange Syndrome[3] "Cornelia de Lange Syndrome (CdLS) is the founding member of a class of multi-organ system birth defect syndromes termed cohesinopathies, named for the chromatin-associated protein complex cohesin, which mediates sister chromatid cohesion. Most cases of CdLS are caused by haploinsufficiency for Nipped-B-like (Nipbl), a highly conserved protein that facilitates cohesin loading. ... These findings support the view that birth defects in CdLS arise from collective effects of quantitative changes in gene expression. Interestingly, both the phenotypes and gene expression changes in nipbl morphants differed from those in mutants or morphants for genes encoding cohesin subunits, suggesting that the transcriptional functions of Nipbl cannot be ascribed simply to its role in cohesin loading. (OMIM - CDLS1 | CDLS2 | CDLS3)
- The zebrafish transcriptome during early development[16] "The three earliest developmental stages were similar when comparing highly expressed genes, whereas the 50% epiboly stage differed from the other three stages in the identity of highly expressed genes, number of uniquely expressed genes and enrichment of GO molecular functions. Taken together, these observations indicate a major transition in gene regulation and transcriptional activity taking place between the 512-cell and 50% epiboly stages, in accordance with previous studies."
- Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes[17] " Three of them are due to mutations in zebrafish orthologues of FRAS1, FREM1, or FREM2, large basement membrane protein encoding genes that are mutated in mouse bleb mutants and in human patients suffering from Fraser Syndrome, a rare congenital condition characterized by syndactyly and cryptophthalmos. Fin blistering in a fourth group of zebrafish mutants is caused by mutations in Hemicentin1 (Hmcn1), another large extracellular matrix protein the function of which in vertebrates was hitherto unknown. Our mutant and dose-dependent interaction data suggest a potential involvement of Hmcn1 in Fraser complex-dependent basement membrane anchorage. Furthermore, we present biochemical and genetic data suggesting a role for the proprotein convertase FurinA in zebrafish fin development and cell surface shedding of Fras1 and Frem2, thereby allowing proper localization of the proteins within the basement membrane of forming fins."
|
Movies
| Movie of an immobilized zebrafish embryo development from the 1-cell stage to 85 hours post fertilisation (hpf).[18]
<html5media height="300" width="948">File:zebrafish movie01.mp4</html5media>
|
|
|
Zebrafish Stages
Pharyngula Period
- Transition from Prim 5 to Long-pec
- The body axis begins to straighten and the head straightens out and lifts dorsally
- Notochord is well developed
- Formation of the Dorsal and Ventral Stripe
- Nervous system is hollow and expanding anteriorly
- The brain has developed into 5 distinct lobes
- Seven pharyngeal arch's develop rapidly during this stage
- Pectoral fins begin to develop
- The Circulatory system develops and the heart beats for the first time
- Blood begins to circulate through a closed circuit of channels
- Tactile sensitivity appears and uncoordinated movements occur
Skull
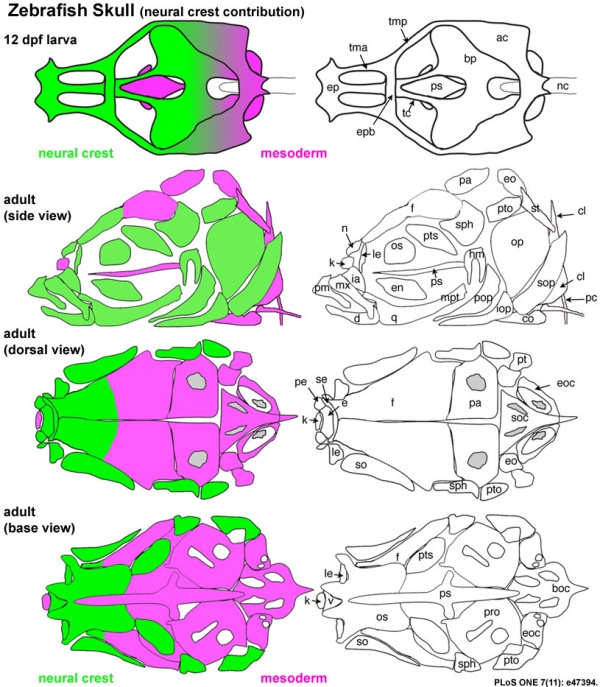
|
Zebrafish Skull Neural Crest Contribution [19]
Diagrams depict the cartilage elements and bones that are NC-derived (green), and those that show no evidence of NC contribution, and are presumably derived from mesoderm (magenta).
- Top - shows a dorsal view of the chondrocranium from an approximately 12 dpf larva.
- Second - side view of the bones of an adult skull, with some elements of the pectoral girdle also shown.
- Third - a dorsal view of the dorsal aspect of the adult skull.
- Bottom - view is of the base of the neurocranium, with the pharyngeal skeleton removed.
(text modified from figure legend)
- Links: Neural Crest Development | Skull Development
|
Neural
Sensory
Lateral line is a zebrafish sensory system, used to detect changes in water flow, composed of clusters of mechanosensory hair cells called neuromasts.
Molecular
Fibroblast Growth Factor
- Fgf8 and Fgf3 - regulating the segmentation of the pharyngeal endoderm into pouches.[20]
- Fgf24 and Fgf8 - promotes posterior mesodermal development.[21]
- Sox9 - required for cartilage morphogenesis.[22]
References
- ↑ Kimmel CB, Sessions SK & Kimmel RJ. (1981). Morphogenesis and synaptogenesis of the zebrafish Mauthner neuron. J. Comp. Neurol. , 198, 101-20. PMID: 7229136 DOI.
- ↑ Kimmel CB, Sepich DS & Trevarrow B. (1988). Development of segmentation in zebrafish. Development , 104 Suppl, 197-207. PMID: 3077108
- ↑ 3.0 3.1 Muto A, Calof AL, Lander AD & Schilling TF. (2011). Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol. , 9, e1001181. PMID: 22039349 DOI.
- ↑ Sharma S, Magnitsky S, Reesey E, Schwartz M, Haroon S, Lavorato M, Chan S, Xiao R, Wilkins BJ, Martinez D, Seiler C & Falk MJ. (2023). Novel Development of Magnetic Resonance Imaging to Quantify the Structural Anatomic Growth of Diverse Organs in Adult and Mutant Zebrafish. Zebrafish , , . PMID: 37603286 DOI.
- ↑ DeLaurier A, Howe DG, Ruzicka L, Carte AN, Mishoe Hernandez L, Wiggins KJ, Gallati MM, Vanpelt K, Loyo Rosado F, Pugh KG, Shabdue CJ, Jihad K, Thyme SB & Talbot JC. (2021). ZebraShare: a new venue for rapid dissemination of zebrafish mutant data. PeerJ , 9, e11007. PMID: 33954026 DOI.
- ↑ Adachi Y, Higuchi A, Wakai E, Shiromizu T, Koiwa J & Nishimura Y. (2021). Involvement of homeobox transcription factor Mohawk in palatogenesis. Congenit Anom (Kyoto) , , . PMID: 34816492 DOI.
- ↑ Schredelseker T & Driever W. (2020). Conserved Genoarchitecture of the Basal Hypothalamus in Zebrafish Embryos. Front Neuroanat , 14, 3. PMID: 32116574 DOI.
- ↑ Li J & Ge W. (2020). Zebrafish as a model for studying ovarian development: Recent advances from targeted gene knockout studies. Mol. Cell. Endocrinol. , 507, 110778. PMID: 32142861 DOI.
- ↑ Nagpal J, Herget U, Choi MK & Ryu S. (2019). Anatomy, development, and plasticity of the neurosecretory hypothalamus in zebrafish. Cell Tissue Res. , 375, 5-22. PMID: 30109407 DOI.
- ↑ Pennisi E. (2018). Development cell by cell. Science , 362, 1344-1345. PMID: 30573610 DOI.
- ↑ Li J, Gao F, Zhao Y, He L, Huang Y, Yang X, Zhou Y, Yu L, Zhao Q & Dong X. (2019). Zebrafish znfl1s regulate left-right asymmetry patterning through controlling the expression of fgfr1a. J. Cell. Physiol. , 234, 1987-1995. PMID: 30317609 DOI.
- ↑ Naylor RW, Qubisi SS & Davidson AJ. (2017). Zebrafish Pronephros Development. Results Probl Cell Differ , 60, 27-53. PMID: 28409341 DOI.
- ↑ Antinucci P & Hindges R. (2016). A crystal-clear zebrafish for in vivo imaging. Sci Rep , 6, 29490. PMID: 27381182 DOI.
- ↑ Xu PF, Houssin N, Ferri-Lagneau KF, Thisse B & Thisse C. (2014). Construction of a vertebrate embryo from two opposing morphogen gradients. Science , 344, 87-9. PMID: 24700857 DOI.
- ↑ Eames BF, DeLaurier A, Ullmann B, Huycke TR, Nichols JT, Dowd J, McFadden M, Sasaki MM & Kimmel CB. (2013). FishFace: interactive atlas of zebrafish craniofacial development at cellular resolution. BMC Dev. Biol. , 13, 23. PMID: 23714426 DOI.
- ↑ Vesterlund L, Jiao H, Unneberg P, Hovatta O & Kere J. (2011). The zebrafish transcriptome during early development. BMC Dev. Biol. , 11, 30. PMID: 21609443 DOI.
- ↑ Carney TJ, Feitosa NM, Sonntag C, Slanchev K, Kluger J, Kiyozumi D, Gebauer JM, Coffin Talbot J, Kimmel CB, Sekiguchi K, Wagener R, Schwarz H, Ingham PW & Hammerschmidt M. (2010). Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet. , 6, e1000907. PMID: 20419147 DOI.
- ↑ Swinburne IA, Mosaliganti KR, Green AA & Megason SG. (2015). Improved Long-Term Imaging of Embryos with Genetically Encoded α-Bungarotoxin. PLoS ONE , 10, e0134005. PMID: 26244658 DOI.
- ↑ Kague E, Gallagher M, Burke S, Parsons M, Franz-Odendaal T & Fisher S. (2012). Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE , 7, e47394. PMID: 23155370 DOI.
- ↑ Crump JG, Maves L, Lawson ND, Weinstein BM & Kimmel CB. (2004). An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development , 131, 5703-16. PMID: 15509770 DOI.
- ↑ Draper BW, Stock DW & Kimmel CB. (2003). Zebrafish fgf24 functions with fgf8 to promote posterior mesodermal development. Development , 130, 4639-54. PMID: 12925590 DOI.
- ↑ Yan YL, Miller CT, Nissen RM, Singer A, Liu D, Kirn A, Draper B, Willoughby J, Morcos PA, Amsterdam A, Chung BC, Westerfield M, Haffter P, Hopkins N, Kimmel C, Postlethwait JH & Nissen R. (2002). A zebrafish sox9 gene required for cartilage morphogenesis. Development , 129, 5065-79. PMID: 12397114
Journals
Zebrafish "is the only peer-reviewed journal to focus on the zebrafish, which has numerous valuable features as a model organism for the study of vertebrate development. Due to its prolific reproduction and the external development of the transparent embryo, the zebrafish is a prime model for genetic and developmental studies, as well as research in toxicology and genomics. While genetically more distant from humans, the vertebrate zebrafish nevertheless has comparable organs and tissues, such as heart, kidney, pancreas, bones, and cartilage." [jour PubMed listing]
Reviews
Supatto W & Vermot J. (2011). From cilia hydrodynamics to zebrafish embryonic development. Curr. Top. Dev. Biol. , 95, 33-66. PMID: 21501748 DOI.
Carvalho L & Heisenberg CP. (2010). The yolk syncytial layer in early zebrafish development. Trends Cell Biol. , 20, 586-92. PMID: 20674361 DOI.
Brittijn SA, Duivesteijn SJ, Belmamoune M, Bertens LF, Bitter W, de Bruijn JD, Champagne DL, Cuppen E, Flik G, Vandenbroucke-Grauls CM, Janssen RA, de Jong IM, de Kloet ER, Kros A, Meijer AH, Metz JR, van der Sar AM, Schaaf MJ, Schulte-Merker S, Spaink HP, Tak PP, Verbeek FJ, Vervoordeldonk MJ, Vonk FJ, Witte F, Yuan H & Richardson MK. (2009). Zebrafish development and regeneration: new tools for biomedical research. Int. J. Dev. Biol. , 53, 835-50. PMID: 19557689 DOI.
Bakkers J, Verhoeven MC & Abdelilah-Seyfried S. (2009). Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev. Biol. , 330, 213-20. PMID: 19371733 DOI.
Chan TM, Longabaugh W, Bolouri H, Chen HL, Tseng WF, Chao CH, Jang TH, Lin YI, Hung SC, Wang HD & Yuh CH. (2009). Developmental gene regulatory networks in the zebrafish embryo. Biochim. Biophys. Acta , 1789, 279-98. PMID: 18992377 DOI.
Articles
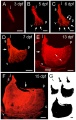
Warga RM & Kane DA. (2018). A Wilson cell origin for Kupffer's vesicle in the zebrafish. Dev. Dyn. , , . PMID: 30016568 DOI.
Search Pubmed
Search Pubmed: Zebrafish Development
Additional Images
Wdr18 expression 6 somite stage
Stages primordial germ cell migration
Non-mammalian VEGF Receptors
Terms
- deep cell layer - (DEL) formed after blastula stage that forms the three germ layers (ectoderm, mesoderm, and endoderm).
- epiboly - (Greek, "epibol" = a throwing or laying on) Term describing the division and movement of ectodermal cells during gastrulation, thinning and spreading this layer to cover the whole of the embryo. Cellular movements are thought to occur in all vertebrates, but have been most clearly identified in both the zebrafish and frog (xenopus laevis).
- enveloping layer - (EVL) an epithelial monolayer formed after blastula stage that undergoes epiboly.
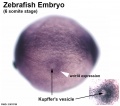
- Kupffer's vesicle - (ciliated organ of asymmetry, primitive node) a transient epithelial fluid-filled sac located midventrally posterior to the yolk cell or its extension. The vesicle has been described as equivalent to the primitive node for establishing embryo left-right (L-R) axis. PMID 21876750 PMID 30016568
- yolk syncytial layer - (YSL) membrane-enclosed group of nuclei that lie on top of the yolk cell formed after blastula stage that undergoes epiboly.
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
Online Atlases
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 25) Embryology Zebrafish Development. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Zebrafish_Development
- What Links Here?
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G