Abnormal Development - Genetic
| Embryology - 19 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
| Educational Use Only - Embryology is an educational resource for learning concepts in embryological development, no clinical information is provided and content should not be used for any other purpose. |
Introduction
This page gives a general introduction to information about genetic abnormalities their relationship to age, ethnicity and prenatal testing. Use the listed links for more information about specific abnormalities. In developed countries with increasing maternal age comes the increased risk of age related genetic abnormalities, such as trisomy 21.
In order to detect some genetic abnormalities many countries offer genetic screening programs that include maternal serum screening (MSS, for detection of Down's syndrome and neural tube defects), embryonic and newborn screening (for phenylketonuria (PKU), hypothyroidism, cystic fibrosis and metabolic disorders).
In terms of maternal/paternal family history, some ethnic backgrounds have been shown to have disease-associated genetic variants, though most common genetic diseases are consistent across ethnic boundaries. For example: Caucasians of northern European ancestry and cystic fibrosis (CTFR gene), Mediterranean, Asian and Far Eastern ancestry with beta-thalassaemia.
Note that the development of in vitro fertilization techniques now allows cells from early stage blastocysts to be removed and genetically analysed prior to implantation. This has raised some ethical issues relating to what parameters will be in future used in blastocyst selection.
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References
|
| Older papers |
|---|
| These papers originally appeared in the Some Recent Findings table, but as that list grew in length have now been shuffled down to this collapsible table.
See also the Discussion Page for other references listed by year and References on this current page.
|
ICD-11 Chromosomal Anomalies
The International Classification of Diseases (ICD) World Health Organization's classification used worldwide as the standard diagnostic tool for epidemiology, health management and clinical purposes.
| ICD-11 Chromosomal anomalies excluding gene mutations | |
|---|---|
| |
Sex chromosome anomalies
| |
| |
| LD7Y Other specified chromosomal anomalies, excluding gene mutations
LD7Z Chromosomal anomalies, excluding gene mutations, unspecified | |
genetic abnormalities
|
Human Chromosomes
| |
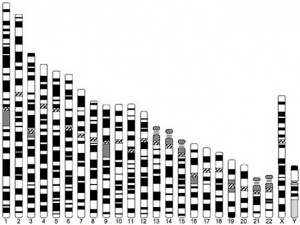
| Idiogram Chromosome Banding - The term refers to the light and dark pattern, seen after staining with a dye, of individual chromosomes identified in metaphase. It is only in meiosis and mitosis during metaphase that chromosomes can be easily identified, during the normal cell life (interphase) the chromosomes are unravelled and distributed within the nucleus in chromosome territories. A band is that part of a chromosome which is clearly distinguishable from nearby regions by appearing darker or brighter with one or more banding techniques. | |
| Genetic abnormality locations: 1-4 | 5-8 | 9-12 | 13-16 | 17-20 | 21-XY | sSMC | |
| |
| Links: Genetics | Abnormal Development - Genetic |
Human Oocyte Aneuploidy
- Frequency and distribution of chromosome abnormalities in human oocytes[11] "It was previously shown that more than half of the human oocytes obtained from IVF patients of advanced reproductive age are aneuploid, due to meiosis I and meiosis II errors. The present paper further confirms that 61.8% of the oocytes tested by fluorescent probes specific for chromosomes 13, 16, 18, 21 and 22 are abnormal, representing predominantly chromatid errors, which are the major source of aneuploidy in the resulting embryos. Almost half of the oocytes with meiosis I errors (49.3%) are prone to sequential meiosis II errors, which may lead to aneuploidy rescue in 30.8% of the cases. Half of the detected aneuploidies (49.8%) are of complex nature with involvement of two or more chromosomes, or the same chromosome in both meiotic divisions. The aneuploidy rates for individual chromosomes are different, with a higher prevalence of chromosome 21 and 22 errors. The origin of aneuploidy for the individual chromosomes is also not random, with chromosome 16 and 22 errors originating more frequently in meiosis II, and chromosome 18, 13 and 21 errors in meiosis I. There is an age dependence not only for the overall frequency of aneuploidies, but also for each chromosome error, aneuploidies originating from meiosis I, meiosis II, and both meiosis I and meiosis II errors, as well as for different types of aneuploidies. The data further suggest the practical relevance of oocyte aneuploidy testing for detection and avoidance from transfer of the embryos deriving from aneuploid oocytes, which should contribute significantly to the pregnancy outcomes of IVF patients of advanced reproduction age."
- Links: Genetic risk maternal age | Trisomy 21 | Trisomy 18 | Trisomy 13 | Oocyte Development
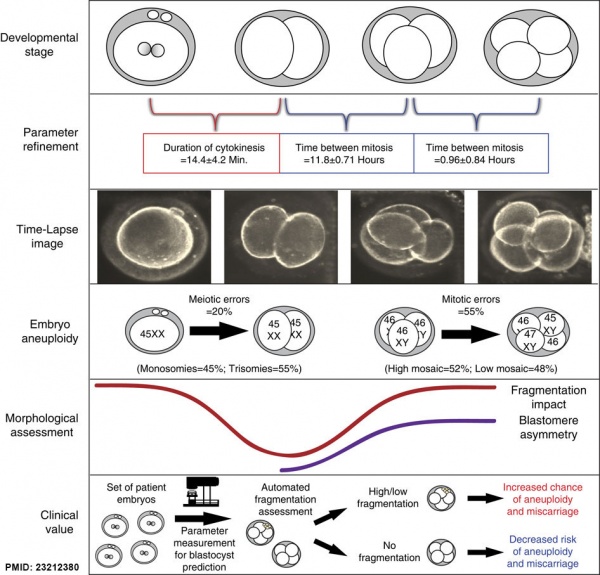
Embryonic Aneuploidy
Embryonic aneuploidy model based on fragmentation.[12] (See also Fragmentation Model and Monosomic Embryo Movie)
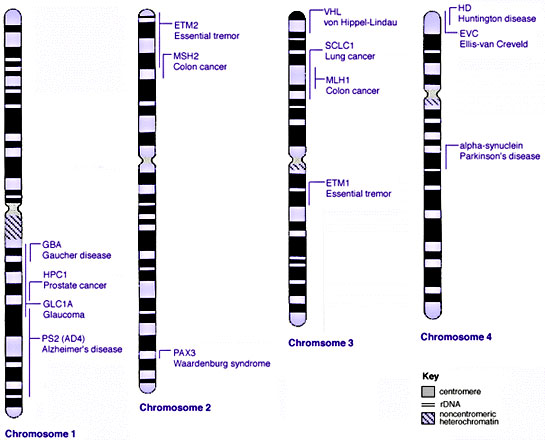
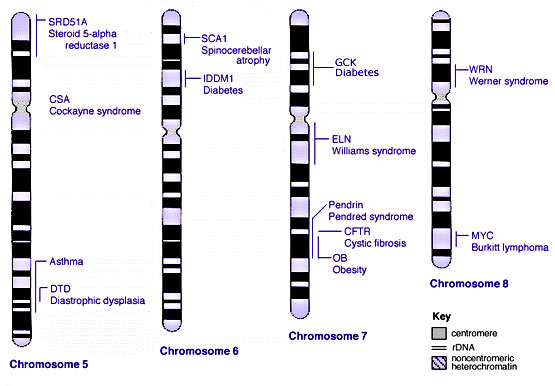
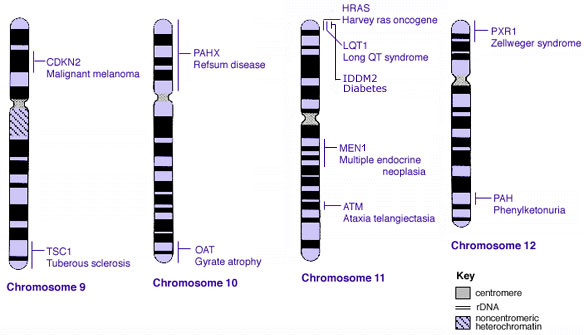
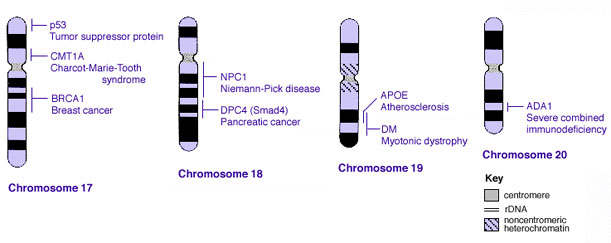
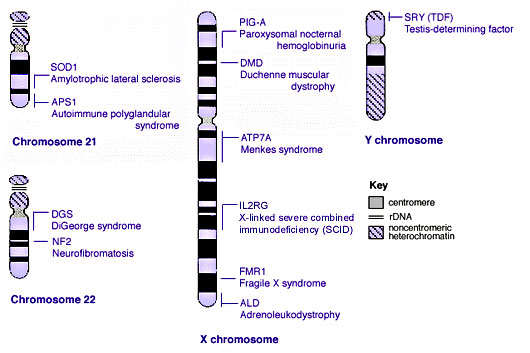
Some Human Disease Gene Locations
Sex Bias
Some developmental abnormalities show a difference in distribution related to embryo sex, Male and Female. While other genetic abnormalities are related to the maternal mitochondrial inheritance sex chromosomes {{ChrX)} and {{ChrY).
Some examples of some abnormalities sex related statistics are shown below.
| Male preponderance | Female preponderance |
|---|---|
|
|
| Cardiac defects | |
|
|
| Table data[7] Links: abnormal development | cardiovascular abnormalities | USA | Male | Female | cleft lip and palate | |
Some neural abnormalities, such as autism and intellectual disability, have a Male sex bias. This can be affected by the presence of specific comorbidities, specific copy number variants, mutational burden, and pre-existing family history of neurodevelopmental phenotypes.[13]
Neural tube defects: Sex ratio changes after fortification with folic acid[14] "Historically, neural tube defects (NTDs) have predominated in Female infants but the reasons remain unclear. In South America, the pre- folic acid fortification (FAF) rates of NTDs were around 18/10,000 births for females and 12/10,000 births for males, with an estimated sex ratio (male/female) of 0.67.
More than 80% of autoimmune disease predominantly affects Females.[15] A possible contribution was suggested in part due to a preponderance of immune system-related genes (ISRG) on the X chromosome, but a recent study has shown these genes are no more abundant on the X chromosome as compared to autosomal chromosomes.[16]
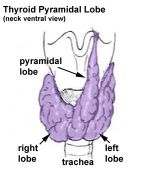
Other abnormalities of development are considered "anatomical variations" as they do not have any serious impacts. For example, the thyroid has a common anatomical variation of a pyramidal lobe seen more frequently in Male than in Female.[17]
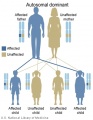
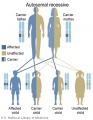
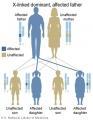
Genetic Inheritance
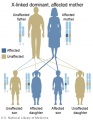
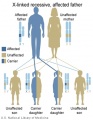
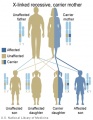
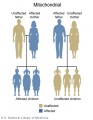
The figures below show the pattern of inheritance of a range of genetic disorders. In addition to these patterns are the known effects of increased maternal age and the effects of genetic mutations in the embryo and newborn.
- Inheritance Pattern images: Genetic Abnormalities | autosomal dominant | autosomal recessive | X-linked dominant (affected father) | X-Linked dominant (affected mother) | X-Linked recessive (affected father) | X-Linked recessive (carrier mother) | mitochondrial inheritance | Codominant inheritance | Genogram symbols | Genetics
An example of autosomal recessive traits are the majority of 50 or so lysosomal diseases, with two X-linked exceptions, that of Fabry disease and Mucopolysaccharidosis II.[18]
Carrier Testing
Preconception carrier tests are available for some autosomal recessive (AR) diseases. These were initially developed for autosomal recessive diseases that were frequent in specific ethnic groups:
- Tay–Sachs disease - Ashkenazi Jews
- hemoglobinopathies - Mediterranean and African populations
- cystic fibrosis - European-derived populations
Genome-wide association study
The National Human Genome Research Institute (USA) lists publications as part of the genome-wide association study (GWAS) in an attempt to assay at least 100,000 single nucleotide polymorphisms (SNPs) in the initial stage.
Fertility
Male Infertility Genes
| Gene abbreviation | Name | Gene Location | Online Mendelian Inheritance in Man (OMIM) |
HUGO Gene Nomenclature Committee (HGNC) |
GeneCards (GCID) | Diagnosis |
|---|---|---|---|---|---|---|
| AURKC | Aurora kinase C | 19q13.43 | 603495 | 11391 | GC19P057230 | Macrozoospermia |
| CATSPER1 | Cation channel sperm-associated 1 | 11q13.1 | 606389 | 17116 | GC11M066034 | Asthenozoospermia |
| CFTR | Cystic fibrosis transmembrane conductance regulator | 7q31.2 | 602421 | 1884 | GC07P117465 | Obstructive azoospermia |
| DNAH1 | Dynein axonemal heavy chain 1 | 3p21.1 | 603332 | 2940 | GC03P052350 | Asthenozoospermia |
| DPY19L2 | Dpy-19-like 2 gene | 12q14.2 | 613893 | 19414 | GC12M063558 | Globozoospermia |
| GALNTL5 | Polypeptide N-acetylgalactosaminyltransferase-like 5 | 7q36.1 | 615133 | 21725 | GC07P151956 | Asthenozoospermia |
| MAGEB4 | MAGE family member B4 | Xp21.2 | 300153 | 6811 | GC0XP030260 | Azoospermia |
| NANOS1 | Nanos C2HC-type zinc finger 1 | 10q26.11 | 608226 | 23044 | GC10P119029 | Azoospermia |
| NR0B1 | Nuclear receptor subfamily 0 group B member 1 | Xp21.2 | 300473 | 7960 | GC0XM030322 | Azoospermia |
| NR5A1 | Nuclear receptor subfamily 5 group A member 1 | 9q33.3 | 184757 | 7983 | GC09M124481 | Azoospermia |
| SOHLH1 | Spermatogenesis and oogenesis-specific basic helix–loop–helix 1 | 9q34.3 | 610224 | 27845 | C09M135693 | Azoospermia |
| vSPATA16 | Spermatogenesis-associated 16 | 3q26.31 | 609856 | 29935 | GC03M172889 | Globozoospermia |
| SYCE1 | Synaptonemal complex central element protein 1 | 10q26.3 | 611486 | 28852 | GC10M133553 | Azoospermia |
| TAF4B | TATA-box binding protein-associated factor 4b | 18q11.2 | 601689 | 11538 | GC18P026225 | Azoospermia |
| TEX11 | Testis expressed 11 | Xq13.1 | 300311 | 11733 | GC0XM070528 | Azoospermia |
| TEX15 | Testis expressed 15, meiosis and synapsis associated | 8p12 | 605795 | 11738 | GC08M030808 | Azoospermia |
| WT1 | Wilms tumour 1 | 8p12 | 607102 | 12796 | GC11M032365 | Azoospermia |
| ZMYND15 | Zinc-finger MYND-type containing 15 | 17p13.2 | 614312 | 20997 | GC17P004740 | Azoospermia |
| Table data source[19] (table 1) Links: fertilization | spermatozoa | testis | Male Infertility Genes | Female Infertility Genes | oocyte | ovary | Genetic Abnormalities | ART Asthenozoospermia - (asthenospermia) term for reduced spermatozoa motility. Azoospermia - term for no spermatozoa located in the ejaculate. Globozoospermia - term for spermatozoa with a round head and no acrosome. | ||||||
Female Infertility Genes
| Gene abbreviation | Name | Gene Location | Online Mendelian Inheritance in Man (OMIM) |
HUGO Gene Nomenclature Committee (HGNC) |
GeneCards (GCID) | Diagnosis |
|---|---|---|---|---|---|---|
| BMP15 | Bone morphogenetic protein 15 | Xp11.22 | 300247 | 1068 | GC0XP050910 | Primary ovarian insufficiency |
| CLPP | Caseinolytic mitochondrial matrix peptidase proteolytic subunit | 19p13.3 | 601119 | 2084 | GC19P006369 | Primary ovarian insufficiency |
| EIF2B2 | Eukaryotic translation initiation factor 2B subunit beta | 14q24.3 | 606454 | 3258 | GC14P075002 | Primary ovarian insufficiency |
| FIGLA | Folliculogenesis-specific BHLH transcription factor | 2p13.3 | 608697 | 24669 | GC02M070741 | Primary ovarian insufficiency |
| FMR1 | Fragile X mental retardation 1 | Xq27.3 | 309550 | 3775 | GC0XP147912 | Primary ovarian insufficiency |
| FOXL2 | Forkhead box L2 | 3q22.3 | 605597 | 1092 | GC03M138944 | Primary ovarian insufficiency |
| FSHR | Follicle stimulating hormone receptor | 2p16.3 | 136435 | 3969 | GC02M048866 | Primary ovarian insufficiency |
| GALT | Galactose-1-phosphate uridylyltransferase | 9p13.3 | 606999 | 4135 | GC09P034636 | Primary ovarian insufficiency |
| GFD9 | Growth differentiation factor 9 | 5q31.1 | 601918 | 4224 | GC05M132861 | Primary ovarian insufficiency |
| HARS2 | Histidyl-TRNA synthetase 2, mitochondrial | 5q31.3 | 600783 | 4817 | GC05P141975 | Primary ovarian insufficiency |
| HFM1 | HFM1, ATP-dependent DNA helicase homolog | 1p22.2 | 615684 | 20193 | GC01M091260 | Primary ovarian insufficiency |
| HSD17B4 | Hydroxysteroid 17-beta dehydrogenase 4 | 5q23.1 | 601860 | 5213 | GC05P119452 | Primary ovarian insufficiency |
| LARS2 | Leucyl-TRNA synthetase 2, mitochondrial | 3p21.31 | 604544 | 17095 | GC03P045405 | Primary ovarian insufficiency |
| LHCGR | Luteinizing hormone/choriogonadotropin receptor | 2p16.3 | 152790 | 6585 | GC02M048647 | Primary ovarian insufficiency |
| LHX8 | LIM homeobox 8 | 1p31.1 | 604425 | 28838 | GC01P075128 | Primary ovarian insufficiency |
| MCM8 | Minichromosome maintenance 8 homologous recombination repair factor | 20p12.3 | 608187 | 16147 | GC20P005926 | Primary ovarian insufficiency |
| MCM9 | Minichromosome maintenance 9 homologous recombination repair factor | 6q22.31 | 610098 | 21484 | GC06M118813 | Primary ovarian insufficiency |
| NOBOX | NOBOX oogenesis homeobox | 7q35 | 610934 | 22448 | GC07M144397 | Primary ovarian insufficiency |
| NOG | Noggin | 17q22 | 602991 | 7866 | GC17P056593 | Primary ovarian insufficiency |
| PMM2 | Phosphomannomutase 2 | 16p13.2 | 601785 | 9115 | GC16P008788 | Primary ovarian insufficiency |
| POLG | DNA polymerase gamma, catalytic subunit | 15q26.1 | 174763 | 9179 | GC15M089316 | Primary ovarian insufficiency |
| REC8 | REC8 meiotic recombination protein | 14q12 | 608193 | 16879 | GC14P024171 | Primary ovarian insufficiency |
| SMC1B | Structural maintenance of chromosomes 1B | 22q13.31 | 608685 | 11112 | GC22M045344 | Primary ovarian insufficiency |
| SOHLH1 | Spermatogenesis and oogenesis-specific basic helix–loop–helix 1 | 9q34.3 | 610224 | 27845 | GC09M135693 | Primary ovarian insufficiency |
| STAG3 | Stromal antigen 3 | 7q22.1 | {{Chr 608489 | 11356 | GC07P100177 | Primary ovarian insufficiency |
| SYCE1 | Synaptonemal Complex Central Element Protein 1 | 10q26.3 | 611486 | 28852 | GC10M133553 | Primary ovarian insufficiency |
| TLE6 | Transducin-like enhancer of split 6 | 19p13.3 | 612399 | 30788 | GC19P002976 | Embryonic lethalithy |
| TUBB8 | Tubulin beta 8 Class VIII | 10p15.3 | 616768 | 20773 | GC10M000048 | Oocyte maturation arrest |
| TWNK | Twinkle MtDNA helicase | 10q24.31 | 606075 | 1160 | GC10P100991 | Primary ovarian insufficiency |
| Table data source[19] (table 1) Links: fertilization | oocyte | ovary | | Female Infertility Genes | spermatozoa | testis | Male Infertility Genes | Genetic Abnormalities | ART Primary ovarian insufficiency - depletion or dysfunction of ovarian follicles with cessation of menses before age 40 years. | ||||||
Genetic Risk Maternal Age
Table Data[20][21][22]
| |||
Uniparental Disomy
Uniparental disomy (UPD) occurs when a person receives two copies of a chromosome, or part of a chromosome, from one parent and no copies from the other parent. This can occur as a random event during the formation of the germ cells (oocyte or spermatozoa) or can also occur in early fetal development.
Trisomy
The term trisomy refers to the abnormal copy number of a specific chromosome, that is 3 copies instead of 2. The abnormality is identified by the chromosome that is present as 3 copies within the cell.
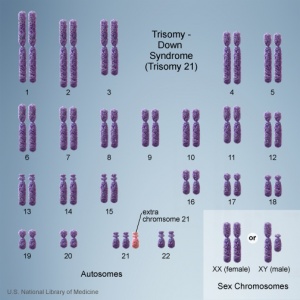
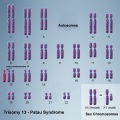
In humans, the most common trisomy is Trisomy 21 or Down syndrome. Other identified human trisomies include Trisomy 13, Trisomy 18 and Trisomy X. Trisomies for other chromosomes are mainly placental in origin and can be associated with miscarriage and growth restriction, or with true fetal mosaicism.
Trisomy 21 (male)
Trisomy 21 (female)
Trisomy Mosaicisms
There have also been a number of trisomy mosaicisms identified for the above and other chromosomes. A "mosaicism" is a rare chromosome disorder characterized by having an extra copy of a chromosome in a proportion, but not all, of a person’s cells.
Identified human examples of this disorder include: Trisomy 22 mosaicism, Trisomy 18 mosaicism, Trisomy 17 mosaicism, Trisomy 14 mosaicism, Trisomy 12 mosaicism, Trisomy 9 mosaicism, Trisomy 8 mosaicism, and Trisomy 2 mosaicism.
- Links: trisomy mosaicism
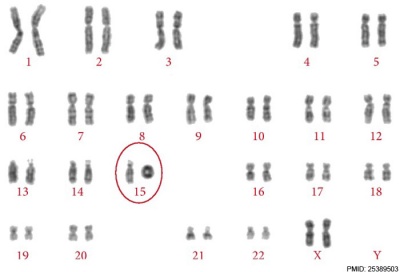
Ring Chromosome
Ring chromosomal 15
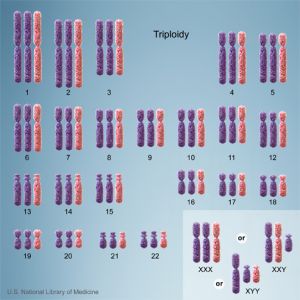
Triploidy
Cells with one additional set of chromosomes, for a total of 69 chromosomes, are called triploid.
|
ICD-11 LD42.0 Triploidy - A disease caused by one additional set of chromosomes, for a total of 69 chromosomes. Triploidy can present with albuminuria, edema, or hypertension in the mother. The fetus may present with microcephaly and a placenta that is enlarged and filled with cysts in the case of extra maternally inherited chromosomes, while extra paternally inherited chromosomes cause severe growth problems, an enlarged head, and a small placenta that does not have cysts. Non-mosaic triploidy is highly lethal, and is rarely observed in live births. Confirmation is through observation of an additional set of chromosomes by karyotyping. |
Tetraploidy
Cells with two additional sets of chromosomes, for a total of 92 chromosomes, are called tetraploid. A condition in which every cell in the body has an extra set of chromosomes and is not compatible with life. Until 1990 there had been been seven reported total number of liveborn infants with a 92,XXYY karyotype.[23][24] Postnatally, tetraploidy can also occur prior to tumorigenesis through nondisjunction of chromosomes in mitosis.[25]
|
ICD-111705084192 LD42.1 Tetraploidy] - A disease caused by two additional sets of chromosomes, for a total of 92 chromosomes. This disease commonly results in spontaneous abortion during the first trimester. Live births of tetraploidy individuals are very rare. These cases are characterized by facial dysmorphism, severely delayed growth and developmental delay. Confirmation is through observation of two additional set of chromosomes by karyotyping. |
Selected Topics
Mutagen
A chemical or agent that can cause permanent damage to the genetic material, DeoxyriboNucleic Acid (DNA) in a cell. DNA damage in the human oocyte or spermatozoa may lead to reduced fertility, spontaneous abortion (miscarriage), birth defects and heritable diseases. DNA damage postnatally can lead to a variety of tumours.
There are a large number of known mutagenic agents including those that are physical, chemical or biological in origin.
Neonatal Testing
The main neonatal genetic test is the Bloodspot Test also known as the Guthrie Test.
Bloodspot Test or Guthrie Test
A blood screening test developed by Dr Robert Guthrie (1916-95) at University of Buffalo.[26] The test is carried out on neonatal (newborn) blood detecting markers for a variety of known disorders (phenylketonuria (PKU), hypothyroidism and cystic fibrosis).
- Links: Guthrie test | neonatal diagnosis
Australia - NSW Newborn Screening Programme
Each year test more than 90,000 babies and detects about 90 who need urgent assessment and treatment. In NSW and Victoria, the bloodspot cards are currently stored indefinitely.
- Phenylketonuria (PKU) - 1 in 10,000 live births (about 10 babies per year). PKU causes high blood levels of phenylalanine and severe intellectual disability. A diet low in phenylalanine, started in the first two to three weeks results in normal development.
- Primary congenital hypothyroidism - 1 in 3,500 live births (about 26 babies per year). It is caused by the absence or abnormal formation or function of the thyroid gland. This causes growth and intellectual disability if not treated. Medication with thyroid hormone started early, results in normal growth and development.
- Cystic Fibrosis (CF) - 1 in 2,500 live births (about 34 babies per year). Without treatment babies develop chest infections and often have very serious failure to thrive. Early institution of treatment greatly improves the health of babies with CF. Newborn bloodspot screening detects about 95% of babies with CF but also detects a few babies who may only be healthy carriers. For these babies a sweat test at about six weeks of age determines whether the baby has CF or is a healthy carrier.
- Galactosaemia - 1 in 40,000 births (about 1-3 cases per year). Babies cannot process galactose, a component of lactose. Life-threatening liver failure and infections can occur. A galactose-free diet instituted in the first week is life saving.
- Rarer metabolic disorders - Some fatty acid, organic acid and other amino acid defects can now be detected using Tandem Mass Spectrometry. These much rarer metabolic disorders affect about 15 – 18 babies per year. Early detection is important as diet and medications can treat most of these disorders. Without appropriate management they can cause severe disability or death.
Genetics services in NSW - coordinated by the NSW Genetics Service Advisory Committee, which is supported by the Statewide Services Development Branch of the Strategic Development Division, NSW Department of Health. (Information from NSW Health - Newborn Bloodspot Screening Policy 13-Nov-2006)
neonatal diagnosis | NSW Genetics Health
Science Student Projects 2011
The links below are to pages prepared as part of an undergraduate science student projects carried out in the Embryology course. Please note these are student submissions and may include inaccuracies in either description or understanding.
- 2011 Projects: Turner Syndrome | DiGeorge Syndrome | Klinefelter's Syndrome | Huntington's Disease | Fragile X Syndrome | Tetralogy of Fallot | Angelman Syndrome | Friedreich's Ataxia | Williams-Beuren Syndrome | Duchenne Muscular Dystrolphy | Cleft Palate and Lip
International Classification of Diseases
Some examples of genetic abnormalities from the ICD-11 classification system.
| ICD-11 |
|---|
|
ICD-11 - LD40.0 Complete trisomy 21 - a chromosomal abnormality, characterised by the presence of a third (partial or total) copy of chromosome 21, which clinical manifestations include variable intellectual deficiency, muscular hypotonia and joint laxity, often associated with facial dysmorphism and variable malformations (essentially heart and digestive) and a risk of complications (epilepsy, leukemia, auto-immune and endocrine pathologies, earlier aging and Alzheimer disease. LD40.1 Complete trisomy 13 - a chromosomal anomaly caused by the presence of an extra chromosome 13 and is characterized by brain malformations (holoprosencephaly), facial dysmorphism, ocular anomalies, postaxial polydactyly, visceral malformations (cardiopathy) and severe psychomotor retardation. LD40.2 Complete trisomy 18 - a chromosomal abnormality associated with the presence of an extra chromosome 18 and characterized by growth delay, dolichocephaly, a characteristic facies, limb anomalies and visceral malformations. LD42.0 Triploidy - A disease caused by one additional set of chromosomes, for a total of 69 chromosomes. Triploidy can present with albuminuria, edema, or hypertension in the mother. The fetus may present with microcephaly and a placenta that is enlarged and filled with cysts in the case of extra maternally inherited chromosomes, while extra paternally inherited chromosomes cause severe growth problems, an enlarged head, and a small placenta that does not have cysts. Non-mosaic triploidy is highly lethal, and is rarely observed in live births. Confirmation is through observation of an additional set of chromosomes by karyotyping. LD42.1 Tetraploidy - A disease caused by two additional sets of chromosomes, for a total of 92 chromosomes. This disease commonly results in spontaneous abortion during the first trimester. Live births of tetraploidy individuals are very rare. These cases are characterized by facial dysmorphism, severely delayed growth and developmental delay. Confirmation is through observation of two additional set of chromosomes by karyotyping. LD45 Uniparental disomies - Any disease caused by the inheritance of two homologous copies of a chromosome from one parent, and none from the other parent. Confirmation is by observation of identical chromosomes pairs by genetic testing. LD50.0 Turner syndrome - Karyotype missing one X chromosome (45, X0 or 45,XO/46,XX mosaicism) ; gonads: ovaries (streak); phenotype female with short stature, amenorrhea (hypergonadotrophic hypogonadism), absence of sexual development, webbed neck, low set ears, posterior hairline, widely-spaced nipples, short fourth metacarpals, and increased carrying angle at the elbow (cubitus valgus). Often associated with renal, cardiac and ocular abnormalities. LD50.1 Karyotype 47,XXX - Trisomy X is a sex chromosome anomaly with a variable phenotype caused by the presence of an extra X chromosome in females (47,XXX instead of 46,XX). Most individuals are only mildly affected or asymptomatic, the most common physical features including tall stature, epicanthal folds, hypotonia and clinodactyly, with seizures, renal and genitourinary abnormalities, and premature ovarian failure being also associated findings. LD50.3 Klinefelter syndrome - Klinefelter syndrome defines a group of chromosomal disorders in which there is at least one extra X chromosome compared with the normal 46,XY male karyotype. The effects on physical features and on physical and cognitive development increase with the number of extra X's, and each extra X is associated with an intelligence quotient (IQ) decrease of approximately 15-16 points, with language most affected, particularly expressive language skills. LD50.30 Klinefelter syndrome with karyotype 47,XXY, regular - Karyotype 47 XXY; gonads: testes (hypogonadism) small and firm with decreased spermatogenesis ; phenotype male with associated congenital abnormalities (decreased virilization due to decreased testosterone production, long arms and legs, short trunk, psychosocial problems) LD50.31 Klinefelter syndrome, male with more than two X chromosomes - A disease affecting males, caused by the presence of more than two X chromosomes in each cell. This disease is characterized by impaired sexual development, intellectual disability, distinctive facial features, skeletal abnormalities, poor coordination, and severe problems with speech. This disease may be differentiated from classic Klinefelter syndrome by increased severity of symptoms. Confirmation is through observation of more than two X chromosomes by karyotyping. LD55 Fragile X chromosome - Fragile X syndrome is a rare genetic disease associated with mild to severe intellectual deficit that may be associated with behavioral disorders and characteristic physical features. Prevalence is estimated at approximately 1/ 2,500 (prevalence of the full mutation allele) to 1/ 4,000 (prevalence of symptomatic cases) for both genders. JA02.0 Complete hydatidiform mole - A condition caused by the over-production of cells arising into the placenta during pregnancy. This condition is characterized by a pregnancy with abnormal placental growth in which the chorionic villi become hydropic, slight to severe trophoblast proliferation and invasion of the uterine tissue within 10-16 weeks after conception, a placental mass, 25-30% theca lutein cysts, 15-20% persistent trophoblastic disease, 50% uterine size for dates, and vaginal bleeding, nausea, or vomiting. This condition leads to an absent fetus. JA02.1 Incomplete or partial hydatidiform mole - A condition caused by the over-production of cells arising into the placenta during pregnancy. This condition is characterized by a pregnancy with abnormal placental growth in which the chorionic villi become hydropic, slight to moderate trophoblast proliferation and invasion of the uterine tissue within 10-16 weeks after conception, a placental mass, theca lutein cysts, 1-5% persistent trophoblastic disease, small uterine size for dates, and vaginal bleeding, nausea, or vomiting. This condition leads to some fetal development and a missed abortion. |
References
- ↑ Elias D, Campaña H, Poletta F, Heisecke S, Gili J, Ratowiecki J, Gimenez L, Pawluk M, Santos MR, Cosentino V, Uranga R, Rittler M & Lopez Camelo J. (2020). A graph theory approach to analyze birth defect associations. PLoS ONE , 15, e0233529. PMID: 32442191 DOI.
- ↑ Bartal MF, Sibai BM, Bart Y, Shina A, Mazaki-Tovi S, Eisen IS, Hendler I, Baum M & Schiff E. (2018). The Impact of Sperm and Egg Donation on the Risk of Pregnancy Complications. Am J Perinatol , , . PMID: 30031370 DOI.
- ↑ Arya S, Mulla ZD & Plavsic SK. (2018). Outcomes of Women Delivering at Very Advanced Maternal Age. J Womens Health (Larchmt) , , . PMID: 30016194 DOI.
- ↑ Wang S, Hassold T, Hunt P, White MA, Zickler D, Kleckner N & Zhang L. (2017). Inefficient Crossover Maturation Underlies Elevated Aneuploidy in Human Female Meiosis. Cell , 168, 977-989.e17. PMID: 28262352 DOI.
- ↑ Jaruthamsophon K, Sriplung H, Charalsawadi C & Limprasert P. (2016). Maternal Age-Specific Rates for Trisomy 21 and Common Autosomal Trisomies in Fetuses from a Single Diagnostic Center in Thailand. PLoS ONE , 11, e0165859. PMID: 27812158 DOI.
- ↑ De Wert G, Heindryckx B, Pennings G, Clarke A, Eichenlaub-Ritter U, van El CG, Forzano F, Goddijn M, Howard HC, Radojkovic D, Rial-Sebbag E, Dondorp W, Tarlatzis BC & Cornel MC. (2018). Responsible innovation in human germline gene editing: Background document to the recommendations of ESHG and ESHRE. Eur. J. Hum. Genet. , , . PMID: 29326429 DOI.
- ↑ 7.0 7.1 Michalski AM, Richardson SD, Browne ML, Carmichael SL, Canfield MA, VanZutphen AR, Anderka MT, Marshall EG & Druschel CM. (2015). Sex ratios among infants with birth defects, National Birth Defects Prevention Study, 1997-2009. Am. J. Med. Genet. A , 167A, 1071-81. PMID: 25711982 DOI.
- ↑ Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL & Ledbetter DH. (2010). Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. , 86, 749-64. PMID: 20466091 DOI.
- ↑ Jugessur A, Shi M, Gjessing HK, Lie RT, Wilcox AJ, Weinberg CR, Christensen K, Boyles AL, Daack-Hirsch S, Nguyen TT, Christiansen L, Lidral AC & Murray JC. (2010). Maternal genes and facial clefts in offspring: a comprehensive search for genetic associations in two population-based cleft studies from Scandinavia. PLoS ONE , 5, e11493. PMID: 20634891 DOI.
- ↑ Jefferson A, Colella S, Moralli D, Wilson N, Yusuf M, Gimelli G, Ragoussis J & Volpi EV. (2010). Altered intra-nuclear organisation of heterochromatin and genes in ICF syndrome. PLoS ONE , 5, e11364. PMID: 20613881 DOI.
- ↑ Kuliev A, Cieslak J & Verlinsky Y. (2005). Frequency and distribution of chromosome abnormalities in human oocytes. Cytogenet. Genome Res. , 111, 193-8. PMID: 16192694 DOI.
- ↑ Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B & Reijo Pera RA. (2012). Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun , 3, 1251. PMID: 23212380 DOI.
- ↑ Polyak A, Rosenfeld JA & Girirajan S. (2015). An assessment of sex bias in neurodevelopmental disorders. Genome Med , 7, 94. PMID: 26307204 DOI.
- ↑ Poletta FA, Rittler M, Saleme C, Campaña H, Gili JA, Pawluk MS, Gimenez LG, Cosentino VR, Castilla EE & López-Camelo JS. (2018). Neural tube defects: Sex ratio changes after fortification with folic acid. PLoS ONE , 13, e0193127. PMID: 29538416 DOI.
- ↑ Liu K, Kurien BT, Zimmerman SL, Kaufman KM, Taft DH, Kottyan LC, Lazaro S, Weaver CA, Ice JA, Adler AJ, Chodosh J, Radfar L, Rasmussen A, Stone DU, Lewis DM, Li S, Koelsch KA, Igoe A, Talsania M, Kumar J, Maier-Moore JS, Harris VM, Gopalakrishnan R, Jonsson R, Lessard JA, Lu X, Gottenberg JE, Anaya JM, Cunninghame-Graham DS, Huang AJW, Brennan MT, Hughes P, Illei GG, Miceli-Richard C, Keystone EC, Bykerk VP, Hirschfield G, Xie G, Ng WF, Nordmark G, Eriksson P, Omdal R, Rhodus NL, Rischmueller M, Rohrer M, Segal BM, Vyse TJ, Wahren-Herlenius M, Witte T, Pons-Estel B, Alarcon-Riquelme ME, Guthridge JM, James JA, Lessard CJ, Kelly JA, Thompson SD, Gaffney PM, Montgomery CG, Edberg JC, Kimberly RP, Alarcón GS, Langefeld CL, Gilkeson GS, Kamen DL, Tsao BP, McCune WJ, Salmon JE, Merrill JT, Weisman MH, Wallace DJ, Utset TO, Bottinger EP, Amos CI, Siminovitch KA, Mariette X, Sivils KL, Harley JB & Scofield RH. (2016). X Chromosome Dose and Sex Bias in Autoimmune Diseases: Increased Prevalence of 47,XXX in Systemic Lupus Erythematosus and Sjögren's Syndrome. , 68, 1290-1300. PMID: 26713507 DOI.
- ↑ Meester I, Manilla-Muñoz E, León-Cachón RBR, Paniagua-Frausto GA, Carrión-Alvarez D, Ruiz-Rodríguez CO, Rodríguez-Rangel X & García-Martínez JM. (2020). SeXY chromosomes and the immune system: reflections after a comparative study. Biol Sex Differ , 11, 3. PMID: 31937374 DOI.
- ↑ Braun EM, Windisch G, Wolf G, Hausleitner L & Anderhuber F. (2007). The pyramidal lobe: clinical anatomy and its importance in thyroid surgery. Surg Radiol Anat , 29, 21-7. PMID: 17146601 DOI.
- ↑ Lee SS, Hadengue A, Girod C, Braillon A & Lebrec D. (1991). Divergent circulatory effects of betaxolol in conscious and anesthetized normal and portal hypertensive rats. J. Hepatol. , 12, 157-61. PMID: 2050994
- ↑ 19.0 19.1 Harper JC, Aittomäki K, Borry P, Cornel MC, de Wert G, Dondorp W, Geraedts J, Gianaroli L, Ketterson K, Liebaers I, Lundin K, Mertes H, Morris M, Pennings G, Sermon K, Spits C, Soini S, van Montfoort APA, Veiga A, Vermeesch JR, Viville S & Macek M. (2018). Recent developments in genetics and medically assisted reproduction: from research to clinical applications. Eur. J. Hum. Genet. , 26, 12-33. PMID: 29199274 DOI.
- ↑ Hook EB. (1981). Rates of chromosome abnormalities at different maternal ages. Obstet Gynecol , 58, 282-5. PMID: 6455611
- ↑ Hook EB, Cross PK & Schreinemachers DM. (1983). Chromosomal abnormality rates at amniocentesis and in live-born infants. JAMA , 249, 2034-8. PMID: 6220164
- ↑ Schreinemachers DM, Cross PK & Hook EB. (1982). Rates of trisomies 21, 18, 13 and other chromosome abnormalities in about 20 000 prenatal studies compared with estimated rates in live births. Hum. Genet. , 61, 318-24. PMID: 6891368
- ↑ López Pajares I, Delicado A, Diaz de Bustamante A, Pellicer A, Pinel I, Pardo M & Martin M. (1990). Tetraploidy in a liveborn infant. J. Med. Genet. , 27, 782-3. PMID: 2074564
- ↑ Scarbrough PR, Hersh J, Kukolich MK, Carroll AJ, Finley SC, Hochberger R, Wilkerson S, Yen FF & Althaus BW. (1984). Tetraploidy: a report of three live-born infants. Am. J. Med. Genet. , 19, 29-37. PMID: 6496571 DOI.
- ↑ Margolis RL. (2005). Tetraploidy and tumor development. Cancer Cell , 8, 353-4. PMID: 16286243 DOI.
- ↑ GUTHRIE R & SUSI A. (1963). A SIMPLE PHENYLALANINE METHOD FOR DETECTING PHENYLKETONURIA IN LARGE POPULATIONS OF NEWBORN INFANTS. Pediatrics , 32, 338-43. PMID: 14063511
Reviews
Webster A & Schuh M. (2017). Mechanisms of Aneuploidy in Human Eggs. Trends Cell Biol. , 27, 55-68. PMID: 27773484 DOI.
Articles
Search Pubmed
Search Pubmed: genetic developmental abnormality
Terms
| Genetic Terms (expand to view) | ||
|---|---|---|
genetic abnormalities | Molecular Development | meiosis | mitosis
| ||
|
| Cell Division Terms (expand to view) | ||
|---|---|---|
meiosis | mitosis
| ||
|
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- Australia NHMRC - Genetics in Family Medicine: The Australian Handbook for General Practitioners Testing and pregnancy PDF (2007)
- Online Mendelian Inheritance in Man
- NHGRI Catalog of Published Genome-Wide Association Studies | PDF
- Genome-Wide Associations (GWA) Karyogram
- Idiogram Album David Adler
- NSW Centre for Genetics Education - Fact Sheets
- The BabySeq Project | Clinical Trials - "The BabySeq Project is a first-of-its-kind randomized clinical trial designed to examine how best to use genomics in clinical pediatric medicine by creating and safely testing methods for integrating sequencing into the care of newborns."
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 19) Embryology Abnormal Development - Genetic. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Abnormal_Development_-_Genetic
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G