| Embryology - 20 Apr 2024 |
|---|
| Google Translate - select your language from the list shown below (this will open a new external page) |
|
العربية | català | 中文 | 中國傳統的 | français | Deutsche | עִברִית | हिंदी | bahasa Indonesia | italiano | 日本語 | 한국어 | မြန်မာ | Pilipino | Polskie | português | ਪੰਜਾਬੀ ਦੇ | Română | русский | Español | Swahili | Svensk | ไทย | Türkçe | اردو | ייִדיש | Tiếng Việt These external translations are automated and may not be accurate. (More? About Translations) |
Introduction

The name derived from Drosophila gene "paired" (prd) with a box (homeodomain) domain. A transcription factor of the helix-turn-helix structural family, DNA binding, and activating gene expression. In human, there are nine member proteins from Pax1 to Pax9.
Pax6 has been identified as regulating development of the central nervous system, eyes, nose, pancreas and pituitary gland.
Developmental Functions: Mesoderm | Neural | Vision | Pancreas | Pituitary | Thymus
| Factor Links: AMH | hCG | BMP | sonic hedgehog | bHLH | HOX | FGF | FOX | Hippo | LIM | Nanog | NGF | Nodal | Notch | PAX | retinoic acid | SIX | Slit2/Robo1 | SOX | TBX | TGF-beta | VEGF | WNT | Category:Molecular |
Some Recent Findings
|
| More recent papers |
|---|
|
This table allows an automated computer search of the external PubMed database using the listed "Search term" text link.
More? References | Discussion Page | Journal Searches | 2019 References | 2020 References Search term: Development Pax <pubmed limit=5>Development Pax</pubmed> |
Human PAX Family
| Table - Human Pax Family | ||||
| Approved Symbol |
Approved Name | Previous Symbols |
Synonyms | Chromosome |
|---|---|---|---|---|
| PAX1 | paired box 1 | 20p11.22 | ||
| PAX2 | paired box 2 | 10q24.31 | ||
| PAX3 | paired box 3 | WS1 | HUP2 | 2q36.1 |
| PAX4 | paired box 4 | MODY9 | 7q32.1 | |
| PAX5 | paired box 5 | BSAP | 9p13.2 | |
| PAX6 | paired box 6 | AN2 | "D11S812E, AN, WAGR" | 11p13 |
| PAX7 | paired box 7 | Hup1 | 1p36.13 | |
| PAX8 | paired box 8 | 2q14.1 | ||
| PAX9 | paired box 9 | 14q13.3 | ||
| Links: Developmental Signals - Pax | OMIM Pax1 | HGNC | Bmp Family | Fgf Family | Pax Family | Sox Family | Tbx Family | ||||
| Human PAX Family | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
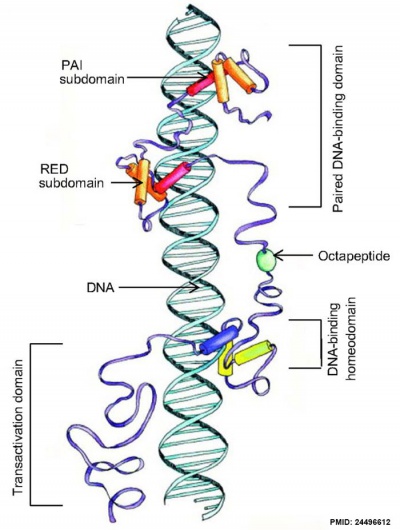
Transcription Factor
Pax and DNA molecular interaction[5]
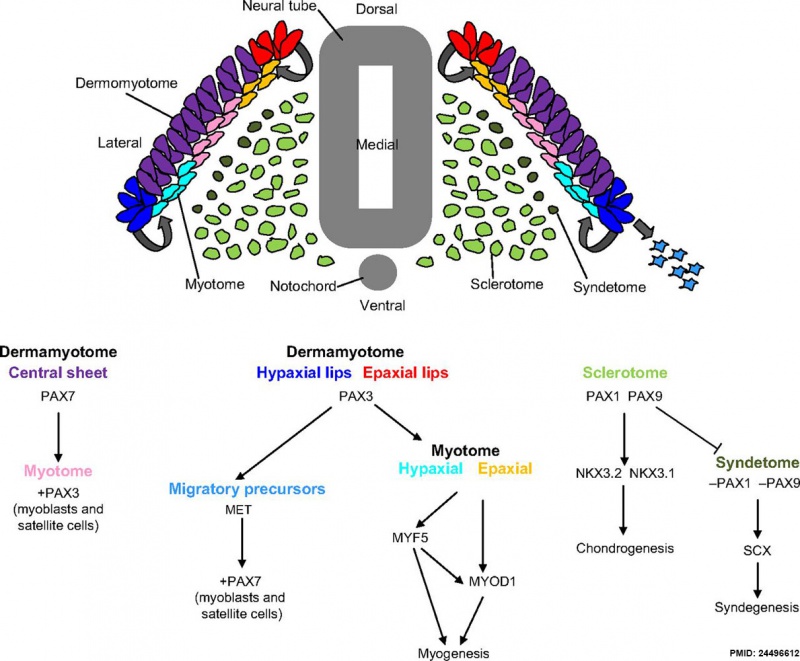
Mesoderm Development
Mesoderm Development and Pax[5]
Neural Development
- Hoxd4 gene a direct target of Pax6[8]
- mouse embryo - Hoxd4 expression in rhombomere 7 and the spinal cord is reduced to some extent in the Pax6 mutant
- zebrafish embryo - double knockdown of pax6a and pax6b with MOs resulted in malformed rhombomere boundaries and an anteriorized hoxd4a expression border
- Pax3 is expressed in the somite, neural tube, and neural crest.
- Pax3 is required for enteric ganglia formation.[9]
- Pax2 and Pax5 in midbrain and cerebellum development.[10]
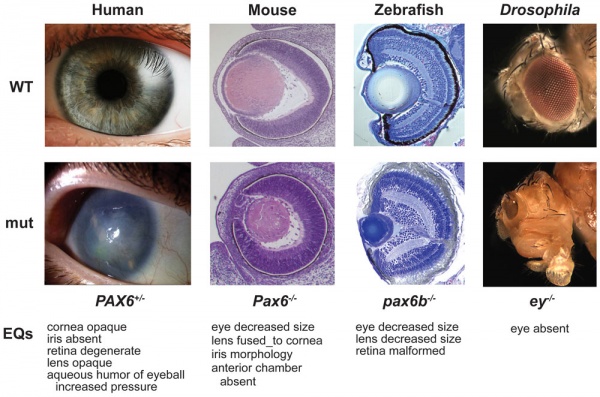
Vision Development
Pax6 mutation eye phenotypes[11]
Pancreas Development
- Pax6 acts in endocrine development in the pancreas as a glucagon gene transactivator role in alpha (α) cell development.
- Pax2 is also expressed in the pancreas.
- Pax4 is a regulator of pancreatic beta cell development.[12]
| Developmental Factors | |
|---|---|
|
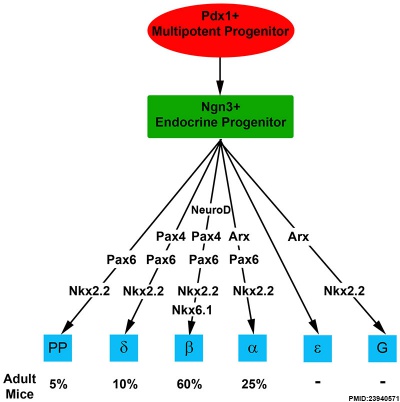
Molecular Development of Endocrine Pancreas Cells[13] |
- Links: Endocrine Pancreas
Thymus Development
Pax1 mouse KO thymus size reduction and impaired thymocyte maturation.
- Links: Thymus Pancreas
Structure
- tissue-specific transcriptional regulators
- contain a highly conserved DNA-binding domain with six alpha-helices (paired domain)
- a complete or residual homeodomain.
- 4 Groups: group I (Pax-1, 9), II (Pax-2, 5, 8), III (Pax-3, 7), and IV (Pax-4, 6)[14]
Mouse Expression
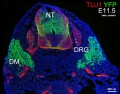
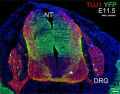
The following gallery is from a recent paper using a Pax7-cre/reporter mouse.[15]
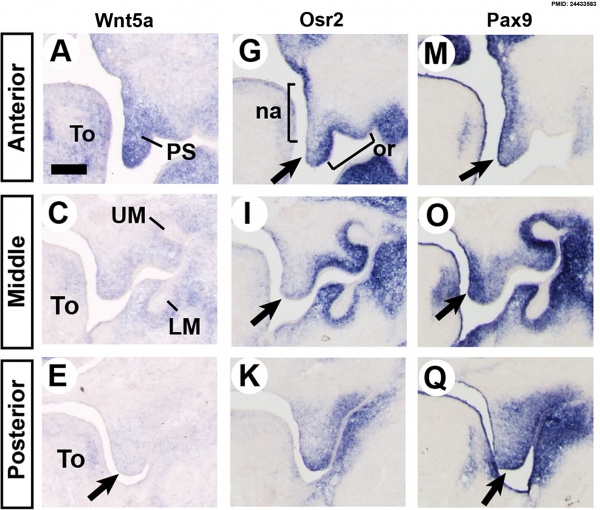
Mouse Palatal Shelf Wnt5a, Osr2 and Pax9 Expression.[16]
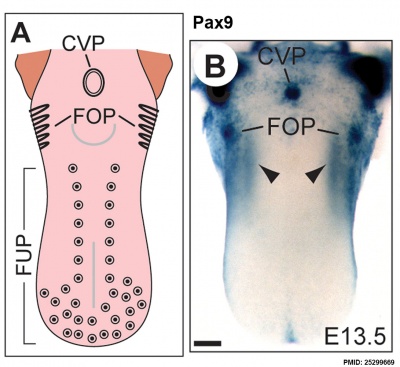
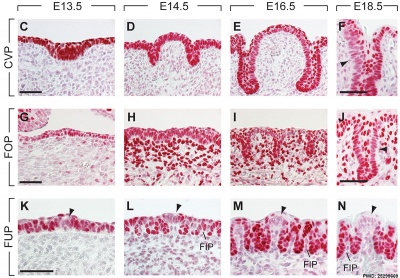
The following data is from a recent paper using a Pax9 reporter.[17]
Abnormalities
Associated with defects in each Pax protein or their signaling pathway.
Pax2
- renal-coloboma syndrome (RCS)
Pax3
- Waardenburg syndrome type 1 (WS1)
- Waardenburg syndrome type 3 (WS3)
- craniofacial-deafness-hand syndrome (CDHS)
- rhabdomyosarcoma type 2 (RMS2)
Pax5
- acute lymphoblastic leukemia
Pax6
A series of vision associated defects.
- aniridia (AN)
- Peters anomaly
- ectopia pupillae
- foveal hypoplasia
- autosomal dominant keratitis
- ocular coloboma
- coloboma of optic nerve
- bilateral optic nerve hypoplasia
- aniridia cerebellar ataxia and mental deficiency (ACAMD)
- Links: Vision Abnormalities
Pax7
- rhabdomyosarcoma type 2 (RMS2)
Pax8
- congenital hypothyroidism non-goitrous type 2 (CHNG2)
- Links:Thyroid Abnormalities
References
- ↑ Sun H, Rodin A, Zhou Y, Dickinson DP, Harper DE, Hewett-Emmett D & Li WH. (1997). Evolution of paired domains: isolation and sequencing of jellyfish and hydra Pax genes related to Pax-5 and Pax-6. Proc. Natl. Acad. Sci. U.S.A. , 94, 5156-61. PMID: 9144207
- ↑ Jia S, Zhou J, Wee Y, Mikkola ML, Schneider P & D'Souza RN. (2017). Anti-EDAR Agonist Antibody Therapy Resolves Palate Defects in Pax9-/- Mice. J. Dent. Res. , 96, 1282-1289. PMID: 28813171 DOI.
- ↑ Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, Winkler S, Hastie AR, Young G, Roscito JG, Falcon F, Knapp D, Powell S, Cruz A, Cao H, Habermann B, Hiller M, Tanaka EM & Myers EW. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature , 554, 50-55. PMID: 29364872 DOI.
- ↑ Navet S, Buresi A, Baratte S, Andouche A, Bonnaud-Ponticelli L & Bassaglia Y. (2017). The Pax gene family: Highlights from cephalopods. PLoS ONE , 12, e0172719. PMID: 28253300 DOI.
- ↑ 5.0 5.1 5.2 Blake JA & Ziman MR. (2014). Pax genes: regulators of lineage specification and progenitor cell maintenance. Development , 141, 737-51. PMID: 24496612 DOI.
- ↑ Numayama-Tsuruta K, Arai Y, Takahashi M, Sasaki-Hoshino M, Funatsu N, Nakamura S & Osumi N. (2010). Downstream genes of Pax6 revealed by comprehensive transcriptome profiling in the developing rat hindbrain. BMC Dev. Biol. , 10, 6. PMID: 20082710 DOI.
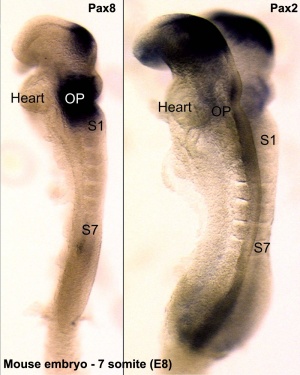
- ↑ Bouchard M, de Caprona D, Busslinger M, Xu P & Fritzsch B. (2010). Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev. Biol. , 10, 89. PMID: 20727173 DOI.
- ↑ <pubmed>17010333</pubmed>
- ↑ Lang D, Chen F, Milewski R, Li J, Lu MM & Epstein JA. (2000). Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J. Clin. Invest. , 106, 963-71. PMID: 11032856 DOI.
- ↑ Schwarz M, Alvarez-Bolado G, Urbánek P, Busslinger M & Gruss P. (1997). Conserved biological function between Pax-2 and Pax-5 in midbrain and cerebellum development: evidence from targeted mutations. Proc. Natl. Acad. Sci. U.S.A. , 94, 14518-23. PMID: 9405645
- ↑ Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M & Lewis SE. (2009). Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. , 7, e1000247. PMID: 19956802 DOI.
- ↑ Sosa-Pineda B. (2004). The gene Pax4 is an essential regulator of pancreatic beta-cell development. Mol. Cells , 18, 289-94. PMID: 15650323
- ↑ Suissa Y, Magenheim J, Stolovich-Rain M, Hija A, Collombat P, Mansouri A, Sussel L, Sosa-Pineda B, McCracken K, Wells JM, Heller RS, Dor Y & Glaser B. (2013). Gastrin: a distinct fate of neurogenin3 positive progenitor cells in the embryonic pancreas. PLoS ONE , 8, e70397. PMID: 23940571 DOI.
- ↑ <pubmed>9254921</pubmed>
- ↑ Murdoch B, DelConte C & García-Castro MI. (2012). Pax7 lineage contributions to the mammalian neural crest. PLoS ONE , 7, e41089. PMID: 22848431 DOI.
- ↑ Almaidhan A, Cesario J, Landin Malt A, Zhao Y, Sharma N, Choi V & Jeong J. (2014). Neural crest-specific deletion of Ldb1 leads to cleft secondary palate with impaired palatal shelf elevation. BMC Dev. Biol. , 14, 3. PMID: 24433583 DOI.
- ↑ Kist R, Watson M, Crosier M, Robinson M, Fuchs J, Reichelt J & Peters H. (2014). The formation of endoderm-derived taste sensory organs requires a Pax9-dependent expansion of embryonic taste bud progenitor cells. PLoS Genet. , 10, e1004709. PMID: 25299669 DOI.
Search Bookshelf Pax
Reviews
Buckingham M & Relaix F. (2007). The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu. Rev. Cell Dev. Biol. , 23, 645-73. PMID: 17506689 DOI.
Mansouri A, Goudreau G & Gruss P. (1999). Pax genes and their role in organogenesis. Cancer Res. , 59, 1707s-1709s; discussion 1709s-1710s. PMID: 10197584
Search Pubmed
Search Pubmed Now: Pax
http://www.ncbi.nlm.nih.gov/omim
External Links
External Links Notice - The dynamic nature of the internet may mean that some of these listed links may no longer function. If the link no longer works search the web with the link text or name. Links to any external commercial sites are provided for information purposes only and should never be considered an endorsement. UNSW Embryology is provided as an educational resource with no clinical information or commercial affiliation.
- OMIM - Pax6
Glossary Links
- Glossary: A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | Numbers | Symbols | Term Link
Cite this page: Hill, M.A. (2024, April 20) Embryology Developmental Signals - Pax. Retrieved from https://embryology.med.unsw.edu.au/embryology/index.php/Developmental_Signals_-_Pax
- © Dr Mark Hill 2024, UNSW Embryology ISBN: 978 0 7334 2609 4 - UNSW CRICOS Provider Code No. 00098G