File:Phylogenetic tree of Pax genes.jpg
From Embryology
Size of this preview: 665 × 600 pixels. Other resolution: 681 × 614 pixels.
Original file (681 × 614 pixels, file size: 41 KB, MIME type: image/jpeg)
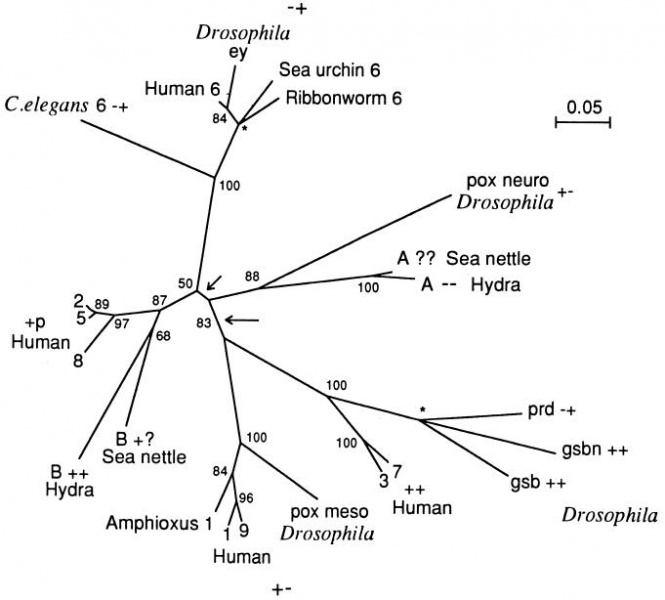
Phylogenetic tree of Pax genes
The tree was inferred from the paired domain sequences (Fig. (Fig.2)2) using Dayhoff’s distances between proteins and the neighbor-joining method.
- The numbers near each branching node denote the bootstrap values. , A trifurcation point.
- The long arrow points to the most plausible root of the tree,
- the short arrow points to an alternative root of the tree.
- The two plus-minus symbols next to a gene name signify the presence (+) or absence (−) of the octapeptide and the paired-type homeodomains, respectively; p, partial; ?, unknown.
Original file name: Figure 5 Pq0970134005.jpg
Reference
<pubmed>9144207</pubmed>
Copyright
Proceedings National Academy of Sciences (PNAS) Liberalization of PNAS copyright policy: Noncommercial use freely allowed Note original Author should be contacted for permission to reuse for Educational purposes. See also PNAS Author Rights and Permission FAQs
- Cozzarelli NR, Fulton KR, Sullenberger DM. Liberalization of PNAS copyright policy: noncommercial use freely allowed. Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12399. PMID15314225 "Our guiding principle is that, while PNAS retains copyright, anyone can make noncommercial use of work in PNAS without asking our permission, provided that the original source is cited."
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 00:49, 23 September 2010 |  | 681 × 614 (41 KB) | S8600021 (talk | contribs) | ==Phylogenetic tree of Pax genes== The tree was inferred from the paired domain sequences (Fig. (Fig.2)2) using Dayhoff’s distances between proteins and the neighbor-joining method. * The numbers near each branching node denote the bootstrap value |
You cannot overwrite this file.
File usage
The following page uses this file: