File:Mouse - blastocoel formation.jpg
Mouse_-_blastocoel_formation.jpg (800 × 319 pixels, file size: 47 KB, MIME type: image/jpeg)
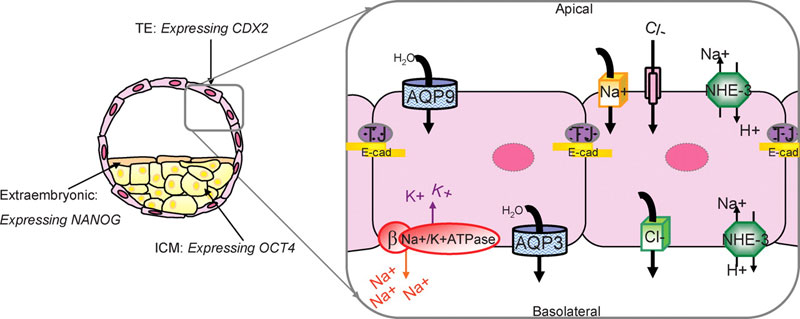
Preimplantation embryo cell lineage specification and blastocyst formation model
Recent research applied to the mouse has established that trophectoderm (TE) and inner cell mass (ICM) differentially express several lineage-specific transcription factors.
- Cdx2 becomes restricted to the TE and is required for TE formation (Yamanaka et al., 2006).
- Oct4 and Nanog become restricted to and influence ICM fate (Yamanaka et al., 2006).
This research supports a model that proposes mutual antagonism between Oct4 and Cdx2 in supporting the formation of TE and ICM fates in the blastocyst (Yamanaka et al., 2006). Our research has been directed at understanding the mechanisms that control blastocyst formation, which is dependent upon TE differentiation, as it is the ion and water transport functions of the TE that mediate the fluid dynamics that control cavitation (Watson et al., 1992; Watson and Barcroft, 2001). Na/K-ATPase, AQP (water channels) and tight junction (TJ) proteins have established roles in coordinating blastocyst formation (Watson et al., 1992; Watson and Barcroft, 2001). Our model predicts that blastocyst formation is dependent upon the polarized distribution of the Na/K-ATP?1?1ase confined to the basolateral membrane domains of the TE. This establishes a trans-TE ion gradient that facilitates movement of water across the epithelium facilitated by the presence of both apical and basolateral AQPs. These events combined with the establishment of a TE tight junctional seal to prevent the loss of fluid out of the embryo through paracellular routes result in the expansion of the embryo and the formation of the blastocyst (Watson and Barcroft, 2001) E-cad, E-cadherin; NHE-3, sodium–hydrogen exchanger 3.
Original file name: Figure 2 F2.large.jpg http://molehr.oxfordjournals.org/content/14/12/691/F2.expansion.html
Paper Abstract
- "Preimplantation development shifts from a maternal to embryonic programme rapidly after fertilization. Although the majority of oogenetic products are lost during the maternal to embryonic transition (MET), several do survive this interval to contribute directly to supporting preimplantation development. Embryonic genome activation (EGA) is characterized by the transient expression of several genes that are necessary for MET, and while EGA represents the first major wave of gene expression, a second mid-preimplantation wave of transcription that supports development to the blastocyst stage has been discovered. The application of genomic approaches has greatly assisted in the discovery of stage specific gene expression patterns and the challenge now is to largely define gene function and regulation during preimplantation development. The basic mechanisms controlling compaction, lineage specification and blastocyst formation are defined. The requirement for embryo culture has revealed plasticity in the developmental programme that may exceed the adaptive capacity of the embryo and has fostered important research directions aimed at alleviating culture-induced changes in embryonic programming. New levels of regulation are emerging and greater insight into the roles played by RNA-binding proteins and miRNAs is required. All of this research is relevant due to the necessity to produce healthy preimplantation embryos for embryo transfer, to ensure that assisted reproductive technologies are applied in the most efficient and safest way possible."
Reference
<pubmed>19043080</pubmed>| Mol Hum Reprod.
Special Issue: Emerging Technologies for the Assessment of Gametes and Embryos - The OMICS
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 10:03, 12 October 2010 | 800 × 319 (47 KB) | S8600021 (talk | contribs) | ==Preimplantation embryo cell lineage specification and blastocyst formation model== Recent research applied to the mouse has established that TE and ICM differentially express several lineage-specific transcription factors. * Cdx2 becomes restricted t |
You cannot overwrite this file.
File usage
The following page uses this file: